Nucleolar stress with and without p53
- PMID: 25482194
- PMCID: PMC4164484
- DOI: 10.4161/nucl.32235
Nucleolar stress with and without p53
Abstract
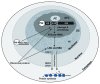
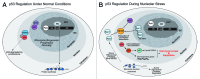
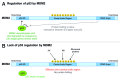
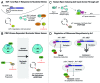
A veritable explosion of primary research papers within the past 10 years focuses on nucleolar and ribosomal stress, and for good reason: with ribosome biosynthesis consuming ~80% of a cell's energy, nearly all metabolic and signaling pathways lead ultimately to or from the nucleolus. We begin by describing p53 activation upon nucleolar stress resulting in cell cycle arrest or apoptosis. The significance of this mechanism cannot be understated, as oncologists are now inducing nucleolar stress strategically in cancer cells as a potential anti-cancer therapy. We also summarize the human ribosomopathies, syndromes in which ribosome biogenesis or function are impaired leading to birth defects or bone narrow failures; the perplexing problem in the ribosomopathies is why only certain cells are affected despite the fact that the causative mutation is systemic. We then describe p53-independent nucleolar stress, first in yeast which lacks p53, and then in other model metazoans that lack MDM2, the critical E3 ubiquitin ligase that normally inactivates p53. Do these presumably ancient p53-independent nucleolar stress pathways remain latent in human cells? If they still exist, can we use them to target >50% of known human cancers that lack functional p53?
Keywords: cell cycle; nucleolar stress; p53; ribosomal proteins; ribosomopathies.
Figures
References
-
- Olson MOJ. Sensing cellular stress: another new function for the nucleolus? Sci STKE. 2004;2004:pe10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
