Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis
- PMID: 25482509
- PMCID: PMC4289414
- DOI: 10.1016/j.molcel.2014.10.028
Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis
Abstract
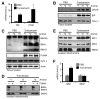
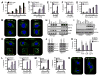
Proapoptotic BCL-2 proteins converge upon the outer mitochondrial membrane (OMM) to promote mitochondrial outer membrane permeabilization (MOMP) and apoptosis. Here we investigated the mechanistic relationship between mitochondrial shape and MOMP and provide evidence that BAX requires a distinct mitochondrial size to induce MOMP. We utilized the terminal unfolded protein response pathway to systematically define proapoptotic BCL-2 protein composition after stress and then directly interrogated their requirement for a productive mitochondrial size. Complementary biochemical, cellular, in vivo, and ex vivo studies reveal that Mfn1, a GTPase involved in mitochondrial fusion, establishes a mitochondrial size that is permissive for proapoptotic BCL-2 family function. Cells with hyperfragmented mitochondria, along with size-restricted OMM model systems, fail to support BAX-dependent membrane association and permeabilization due to an inability to stabilize BAXα9·membrane interactions. This work identifies a mechanistic contribution of mitochondrial size in dictating BAX activation, MOMP, and apoptosis.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Distinct lipid effects on tBid and Bim activation of membrane permeabilization by pro-apoptotic Bax.Biochem J. 2015 May 1;467(3):495-505. doi: 10.1042/BJ20141291. Biochem J. 2015. PMID: 25714678
-
MOMP in the absence of BH3-only proteins.Genes Dev. 2016 Apr 15;30(8):878-80. doi: 10.1101/gad.281519.116. Genes Dev. 2016. PMID: 27083995 Free PMC article.
-
Opa1-mediated cristae opening is Bax/Bak and BH3 dependent, required for apoptosis, and independent of Bak oligomerization.Mol Cell. 2008 Aug 22;31(4):557-569. doi: 10.1016/j.molcel.2008.07.010. Epub 2008 Aug 7. Mol Cell. 2008. PMID: 18691924 Free PMC article.
-
How do BCL-2 proteins induce mitochondrial outer membrane permeabilization?Trends Cell Biol. 2008 Apr;18(4):157-64. doi: 10.1016/j.tcb.2008.01.007. Epub 2008 Mar 7. Trends Cell Biol. 2008. PMID: 18314333 Free PMC article. Review.
-
How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane?Curr Opin Cell Biol. 2006 Dec;18(6):685-9. doi: 10.1016/j.ceb.2006.10.004. Epub 2006 Oct 12. Curr Opin Cell Biol. 2006. PMID: 17046225 Review.
Cited by
-
Mitochondria and Cancer.Cell. 2016 Jul 28;166(3):555-566. doi: 10.1016/j.cell.2016.07.002. Cell. 2016. PMID: 27471965 Free PMC article. Review.
-
A Synthetic Peptide AWRK6 Alleviates Lipopolysaccharide-Induced Liver Injury.Int J Mol Sci. 2018 Sep 7;19(9):2661. doi: 10.3390/ijms19092661. Int J Mol Sci. 2018. PMID: 30205524 Free PMC article.
-
Coenzyme Q10 Modulates Apoptotic Effects of Chronic Administration of Methadone on NMRI Mouse Hippocampus.Cell J. 2021 Oct;23(5):538-543. doi: 10.22074/cellj.2021.7384. Epub 2021 Oct 30. Cell J. 2021. PMID: 34837681 Free PMC article.
-
Dual suppression of inner and outer mitochondrial membrane functions augments apoptotic responses to oncogenic MAPK inhibition.Cell Death Dis. 2018 Jan 18;9(2):29. doi: 10.1038/s41419-017-0044-1. Cell Death Dis. 2018. PMID: 29348439 Free PMC article.
-
miR-340 Inhibits Proliferation and Induces Apoptosis in Gastric Cancer Cell Line SGC-7901, Possibly via the AKT Pathway.Med Sci Monit. 2017 Jan 6;23:71-77. doi: 10.12659/msm.898449. Med Sci Monit. 2017. PMID: 28057912 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

