DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia
- PMID: 25482556
- PMCID: PMC4267494
- DOI: 10.1016/j.celrep.2014.11.004
DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia
Abstract
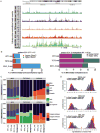
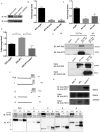
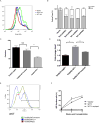
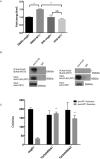
Somatic mutations in IDH1/IDH2 and TET2 result in impaired TET2-mediated conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). The observation that WT1 inactivating mutations anticorrelate with TET2/IDH1/IDH2 mutations in acute myeloid leukemia (AML) led us to hypothesize that WT1 mutations may impact TET2 function. WT1 mutant AML patients have reduced 5hmC levels similar to TET2/IDH1/IDH2 mutant AML. These mutations are characterized by convergent, site-specific alterations in DNA hydroxymethylation, which drive differential gene expression more than alterations in DNA promoter methylation. WT1 overexpression increases global levels of 5hmC, and WT1 silencing reduced 5hmC levels. WT1 physically interacts with TET2 and TET3, and WT1 loss of function results in a similar hematopoietic differentiation phenotype as observed with TET2 deficiency. These data provide a role for WT1 in regulating DNA hydroxymethylation and suggest that TET2 IDH1/IDH2 and WT1 mutations define an AML subtype defined by dysregulated DNA hydroxymethylation.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Abbas S, Erpelinck-Verschueren CAJ, Goudswaard CS, Lowenberg B, Valk PJM. Mutant Wilms' tumor 1 (WT1) mRNA with premature termination codons in acute myeloid leukemia (AML) is sensitive to nonsense-mediated RNA decay (NMD) Leukemia. 2010;24:660–663. - PubMed
-
- Bullinger L, Ehrich M, Dohner K, Schlenk RF, Dohner H, Nelson MR, van den Boom D. Quantitative DNA methylation predicts survival in adult acute myeloid leukemia. Blood. 2010;115:636–642. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 TR000430/TR/NCATS NIH HHS/United States
- U54 CA143798/CA/NCI NIH HHS/United States
- K01 HG006699/HG/NHGRI NIH HHS/United States
- R01 CA166835/CA/NCI NIH HHS/United States
- R01 CA129831/CA/NCI NIH HHS/United States
- U10 CA021115/CA/NCI NIH HHS/United States
- CA129831-03S1/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- P30 CA060553/CA/NCI NIH HHS/United States
- R01 HG006798/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 CA169784/CA/NCI NIH HHS/United States
- R01 CA168802/CA/NCI NIH HHS/United States
- CA172636-01/CA/NCI NIH HHS/United States
- U10 CA180820/CA/NCI NIH HHS/United States
- U24 CA114737/CA/NCI NIH HHS/United States
- R01 CA173636/CA/NCI NIH HHS/United States
- R21 CA176362/CA/NCI NIH HHS/United States
- 01NS076465/NS/NINDS NIH HHS/United States
- R01HG006798/HG/NHGRI NIH HHS/United States
- R01 NS076465/NS/NINDS NIH HHS/United States
- CA129831/CA/NCI NIH HHS/United States
- U54CA143798-01/CA/NCI NIH HHS/United States
- U10 CA180827/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

