Involvement of the S4-S5 linker and the C-linker domain regions to voltage-gating in plant Shaker channels: comparison with animal HCN and Kv channels
- PMID: 25482770
- PMCID: PMC4622754
- DOI: 10.4161/15592316.2014.972892
Involvement of the S4-S5 linker and the C-linker domain regions to voltage-gating in plant Shaker channels: comparison with animal HCN and Kv channels
Abstract
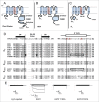
Among the different transport systems present in plant cells, Shaker channels constitute the major pathway for K(+) in the plasma membrane. Plant Shaker channels are members of the 6 transmembrane-1 pore (6TM-1P) cation channel superfamily as the animal Shaker (Kv) and HCN channels. All these channels are voltage-gated K(+) channels: Kv channels are outward-rectifiers, opened at depolarized voltages and HCN channels are inward-rectifiers, opened by membrane hyperpolarization. Among plant Shaker channels, we can find outward-rectifiers, inward-rectifiers and also weak-rectifiers, with weak voltage dependence. Despite the absence of crystal structures of plant Shaker channels, functional analyses coupled to homology modeling, mostly based on Kv and HCN crystals, have permitted the identification of several regions contributing to plant Shaker channel gating. In the present mini-review, we make an update on the voltage-gating mechanism of plant Shaker channels which seem to be comparable to that proposed for HCN channels.
Keywords: C-linker; Shaker; channel; potassium; voltage-gating.
Figures
References
-
- Very AA, Sentenac H. Molecular mechanisms and regulation of K +transport in higher plants. Annu Rev Plant Biol 2003; 54:575-603; PMID:14503004; http://dx.doi.org/10.1146annurev.arplant.54.031902.134831 - PubMed
-
- Lebaudy A, Very AA, Sentenac H. K+ channel activity in plants: genes, regulations and functions. FEBS Lett 2007; 581:2357-66; PMID:17418142; http://dx.doi.org/10.1016j.febslet.2007.03.058 - PubMed
-
- Wang Y, Wu WH. Potassium transport and signaling in higher plants. Annu Rev Plant Biol 2013; 64:451-76; PMID:23330792; http://dx.doi.org/10.1146annurev-arplant-050312-120153 - PubMed
-
- Sharma T, Dreyer I, Riedelsberger J. The role of K(+) channels in uptake and redistribution of potassium in the model plant Arabidopsis thaliana. Front Plant Sci 2013; 4:224; PMID:23818893; http://dx.doi.org/10.3389fpls.2013.00224 - PMC - PubMed
-
- Dreyer I, Blatt MR. What makes a gate? The ins and outs of Kv-like K+ channels in plants. Trends Plant Sci 2009; 14:383-90; PMID:19540150; http://dx.doi.org/10.1016j.tplants.2009.04.001 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
