A tandem affinity purification tag of TGA2 for isolation of interacting proteins in Arabidopsis thaliana
- PMID: 25482810
- PMCID: PMC4622720
- DOI: 10.4161/15592316.2014.972794
A tandem affinity purification tag of TGA2 for isolation of interacting proteins in Arabidopsis thaliana
Abstract
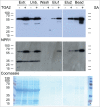
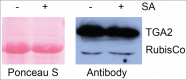
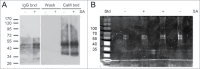
Tandem affinity purification (TAP) tagging provides a powerful tool for isolating interacting proteins in vivo. TAP-tag purification offers particular advantages for the identification of stimulus-induced protein interactions. Type II bZIP transcription factors (TGA2, TGA5 and TGA6) play key roles in pathways that control salicylic acid, ethylene, xenobiotic and reactive oxylipin signaling. Although proteins interacting with these transcription factors have been identified through genetic and yeast 2-hybrid screening, others are still elusive. We have therefore generated a C-terminal TAP-tag of TGA2 to isolate additional proteins that interact with this transcription factor. Three lines most highly expressing TAP-tagged TGA2 were functional in that they partially complemented reactive oxylipin-responsive gene expression in a tga2 tga5 tga6 triple mutant. TAP-tagged TGA2 in the most strongly overexpressing line was proteolytically less stable than in the other 2 lines. Only this overexpressing line could be used in a 2-step purification process, resulting in isolation of co-purifying bands of larger molecular weight than TGA2. TAP-tagged TGA2 was used to pull down NPR1, a protein known to interact with this transcription factor. Mass spectrometry was used to identify peptides that co-purified with TAP-tagged TGA2. Having generated this TGA2 TAP-tag line will therefore be an asset to researchers interested in stimulus-induced signal transduction processes.
Keywords: 12-oxo-phytodienoic acid; CBB, calmodulin binding buffer; CBP, calmodulin-binding peptide; CaMV, cauliflower mosaic virus; FDR, false discovery rate; MS, mass spectrometry; OPDA, 12-oxo-phytodienoic acid; PGA1, prostaglandin A1; PPA1, phytoprostane A1; RubisCo, ribulose-1,5-bisphosphate carboxylase; SA, salicylic acid; SAR, systemic acquired resistance; TAP, tandem affinity purification; TEV, tobacco etch virus; Y2H, yeast 2-hybrid; bZIP, basic region/leucine zipper motif; glutathione-S-transferase; lipid stress; protein complex; thale cress.
Figures



Similar articles
-
Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance.Plant Cell. 2003 Nov;15(11):2647-53. doi: 10.1105/tpc.014894. Epub 2003 Oct 23. Plant Cell. 2003. PMID: 14576289 Free PMC article.
-
TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins.J Exp Bot. 2013 Feb;64(4):963-75. doi: 10.1093/jxb/ers389. Epub 2013 Jan 23. J Exp Bot. 2013. PMID: 23349138 Free PMC article.
-
Over-expression of TGA5, which encodes a bZIP transcription factor that interacts with NIM1/NPR1, confers SAR-independent resistance in Arabidopsis thaliana to Peronospora parasitica.Plant J. 2002 Oct;32(2):151-63. doi: 10.1046/j.1365-313x.2001.01411.x. Plant J. 2002. PMID: 12383081
-
Interaction proteomics: characterization of protein complexes using tandem affinity purification-mass spectrometry.Biochem Soc Trans. 2010 Aug;38(4):883-7. doi: 10.1042/BST0380883. Biochem Soc Trans. 2010. PMID: 20658971 Review.
-
Commonly used tag combinations for tandem affinity purification.Biotechnol Appl Biochem. 2010 Feb 15;55(2):73-83. doi: 10.1042/BA20090273. Biotechnol Appl Biochem. 2010. PMID: 20156193 Review.
Cited by
-
Primary root response to combined drought and heat stress is regulated via salicylic acid metabolism in maize.BMC Plant Biol. 2022 Aug 30;22(1):417. doi: 10.1186/s12870-022-03805-4. BMC Plant Biol. 2022. PMID: 36038847 Free PMC article.
-
Expression Analysis of MaTGA8 Transcription Factor in Banana and Its Defence Functional Analysis by Overexpression in Arabidopsis.Int J Mol Sci. 2021 Aug 28;22(17):9344. doi: 10.3390/ijms22179344. Int J Mol Sci. 2021. PMID: 34502265 Free PMC article.
-
Identification of potential cargo proteins of transportin protein AtTRN1 in Arabidopsis thaliana.Plant Cell Rep. 2016 Mar;35(3):629-40. doi: 10.1007/s00299-015-1908-4. Epub 2015 Dec 9. Plant Cell Rep. 2016. PMID: 26650834
References
-
- Ngounou Wetie AG, Sokolowska I, Woods AG, Roy U, Loo JA, Darie CC. Investigation of stable and transient protein-protein interactions: past, present, and future. Proteomics 2013; 13:538-57; PMID:23193082; http://dx.doi.org/10.1002/pmic.201200328 - DOI - PMC - PubMed
-
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 1999; 17:1030-2; PMID:10504710; http://dx.doi.org/10.1038/13732 - DOI - PubMed
-
- Wang C, Shang JX, Chen QX, Oses-Prieto JA, Bai MY, Yang Y, Yuan M, Zhang YL, Mu CC, Deng Z, et al. . Identification of BZR1-interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification. Mol Cell Proteomics 2013; 12:3653-65; PMID:24019147; http://dx.doi.org/10.1074/mcp.M113.029256 - DOI - PMC - PubMed
-
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Peirats-Llobet M, Pizzio GA, Fernandez MA, De Winne N, De Jaeger G, Dietrich D, Bennett MJ, et al. . PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol 2013; 161:931-41; PMID:23370718; http://dx.doi.org/10.1104/pp.112.208678 - DOI - PMC - PubMed
-
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends Plant Sci 2002; 7:106-11; PMID:11906833; http://dx.doi.org/10.1016/S1360-1385(01)02223-3 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
