mTOR inhibitors blunt the p53 response to nucleolar stress by regulating RPL11 and MDM2 levels
- PMID: 25482947
- PMCID: PMC4623102
- DOI: 10.4161/15384047.2014.955743
mTOR inhibitors blunt the p53 response to nucleolar stress by regulating RPL11 and MDM2 levels
Abstract
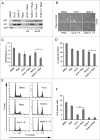
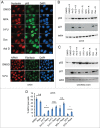
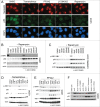
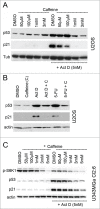
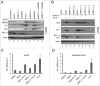
Mechanistic target of rapamycin (mTOR) is a master regulator of cell growth through its ability to stimulate ribosome biogenesis and mRNA translation. In contrast, the p53 tumor suppressor negatively controls cell growth and is activated by a wide range of insults to the cell. The mTOR and p53 signaling pathways are connected by a number of different mechanisms. Chemotherapeutics that inhibit ribosome biogenesis often induce nucleolar stress and activation of p53. Here we have investigated how the p53 response to nucleolar stress is affected by simultaneous mTOR inhibition in osteosarcoma and glioma cell lines. We found that inhibitors of the mTOR pathway including rapamycin, wortmannin, and caffeine blunted the p53 response to nucleolar stress induced by actinomycin D. Synthetic inhibitors of mTOR (temsirolimus, LY294.002 and PP242) also impaired actinomycin D triggered p53 stabilization and induction of p21. Ribosomal protein (RPL11) is known to be required for p53 protein stabilization following nucleolar stress. Treatment of cells with mTOR inhibitors may lead to reduced synthesis of RPL11 and thereby destabilize p53. We found that rapamycin mimicked the effect of RPL11 depletion in terms of blunting the p53 response to nucleolar stress. However, the extent to which the levels of p53 and RPL11 were reduced by rapamycin varied between cell lines. Additional mechanisms whereby rapamycin blunts the p53 response to nucleolar stress are likely to be involved. Indeed, rapamycin increased the levels of endogenous MDM2 despite inhibition of its phosphorylation at Ser-166. Our findings may have implications for the design of combinatorial cancer treatments with mTOR pathway inhibitors.
Keywords: 5-FU, 5-fluorouracil; Act D, actinomycin D; BrdU, bromodeoxyuridine; CHX, cycloheximide; DMSO, dimethylsulphoxide; DOX, doxorubicin; EGCG, epigallocatechin-3-gallate; FACS, fluorescence-activated cell sorting; MPA, mycophenolic acid; MTT, (3-[4, 5-dimethylthiazol-2-yl]-2, 5 diphenyl tetrazolium bromide); PI, propidium iodide; actinomycin D; caffeine; glioma; mTOR; mTOR, mechanistic target of rapamycin; nutlin-3; p21; p53; rapamycin; ribosomal protein L11; ribosome biogenesis.
Figures



References
-
- Bywater MJ, Pearson RB, McArthur GA, Hannan RD. Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat Rev Cancer 2013; 13:299-314; PMID:23612459; http://dx.doi.org/ 10.1038/nrc3496 - DOI - PubMed
-
- Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood 2010; 115:3196-205; PMID:20194897; http://dx.doi.org/ 10.1182/blood-2009-10-178129 - DOI - PMC - PubMed
-
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell 2009; 137:413-31; PMID:19410540; http://dx.doi.org/ 10.1016/j.cell.2009.04.037 - DOI - PubMed
-
- Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nature Rev Cancer 2013; 13:83-96; http://dx.doi.org/ 10.1038/nrc3430 - DOI - PMC - PubMed
-
- Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 2010; 29:4253-60; PMID:20498634; http://dx.doi.org/ 10.1038/onc.2010.189 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
