Genomic analysis of RNA localization
- PMID: 25483039
- PMCID: PMC4615561
- DOI: 10.4161/rna.32146
Genomic analysis of RNA localization
Abstract
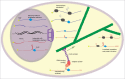
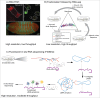
The localization of mRNAs to specific subcellular sites is widespread, allowing cells to spatially restrict and regulate protein production, and playing important roles in development and cellular physiology. This process has been studied in mechanistic detail for several RNAs. However, the generality or specificity of RNA localization systems and mechanisms that impact the many thousands of localized mRNAs has been difficult to assess. In this review, we discuss the current state of the field in determining which RNAs localize, which RNA sequences mediate localization, the protein factors involved, and the biological implications of localization. For each question, we examine prominent systems and techniques that are used to study individual messages, highlight recent genome-wide studies of RNA localization, and discuss the potential for adapting other high-throughput approaches to the study of localization.
Keywords: RNA transport; RNA-seq; fractionation; genomics; subcellular localization; transcriptome; translation.
Figures
Similar articles
-
Why cells move messages: the biological functions of mRNA localization.Semin Cell Dev Biol. 2007 Apr;18(2):171-7. doi: 10.1016/j.semcdb.2007.01.010. Epub 2007 Feb 6. Semin Cell Dev Biol. 2007. PMID: 17398125 Review.
-
Systematic imaging reveals features and changing localization of mRNAs in Drosophila development.Elife. 2015 Apr 2;4:e05003. doi: 10.7554/eLife.05003. Elife. 2015. PMID: 25838129 Free PMC article.
-
Localization of RNAs to the mitochondria-mechanisms and functions.RNA. 2024 May 16;30(6):597-608. doi: 10.1261/rna.079999.124. RNA. 2024. PMID: 38448244 Free PMC article. Review.
-
Genome-Wide Probing of RNA Structures In Vitro Using Nucleases and Deep Sequencing.Methods Mol Biol. 2016;1361:141-60. doi: 10.1007/978-1-4939-3079-1_9. Methods Mol Biol. 2016. PMID: 26483021
-
Classical and emerging techniques to identify and quantify localized RNAs.Wiley Interdiscip Rev RNA. 2019 Sep;10(5):e1542. doi: 10.1002/wrna.1542. Epub 2019 May 1. Wiley Interdiscip Rev RNA. 2019. PMID: 31044542 Review.
Cited by
-
CoLoC-seq probes the global topology of organelle transcriptomes.Nucleic Acids Res. 2023 Feb 22;51(3):e16. doi: 10.1093/nar/gkac1183. Nucleic Acids Res. 2023. PMID: 36537202 Free PMC article.
-
Condensate functionalization with microtubule motors directs their nucleation in space and allows manipulating RNA localization.EMBO J. 2023 Oct 16;42(20):e114106. doi: 10.15252/embj.2023114106. Epub 2023 Sep 19. EMBO J. 2023. PMID: 37724036 Free PMC article.
-
Understanding RNA modifications: the promises and technological bottlenecks of the 'epitranscriptome'.Open Biol. 2017 May;7(5):170077. doi: 10.1098/rsob.170077. Open Biol. 2017. PMID: 28566301 Free PMC article. Review.
-
Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS).Nat Biotechnol. 2019 Feb;37(2):169-178. doi: 10.1038/s41587-018-0001-2. Epub 2019 Jan 3. Nat Biotechnol. 2019. PMID: 30607034 Free PMC article.
-
Neuronal messenger ribonucleoprotein transport follows an aging Lévy walk.Nat Commun. 2018 Jan 24;9(1):344. doi: 10.1038/s41467-017-02700-z. Nat Commun. 2018. PMID: 29367597 Free PMC article.
References
-
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. . The developmental transcriptome of Drosophila melanogaster. Nature 2011; 471:473-9; PMID:21179090; http://dx.doi.org/10.1038/nature09715 - DOI - PMC - PubMed
-
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012; 336:233-7; PMID:22422859; http://dx.doi.org/10.1126/science.1215704 - DOI - PMC - PubMed
-
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012; 484:339-44; PMID:22456710; http://dx.doi.org/10.1038/nature10960 - DOI - PMC - PubMed
-
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 1991; 66:37-50; PMID:2070417; http://dx.doi.org/10.1016/0092-8674(91)90137-N - DOI - PubMed
-
- Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature 1992; 358:387-92; PMID:1641021; http://dx.doi.org/10.1038/358387a0 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
