Mir-23a and mir-125b regulate neural stem/progenitor cell proliferation by targeting Musashi1
- PMID: 25483045
- PMCID: PMC4615800
- DOI: 10.4161/rna.35508
Mir-23a and mir-125b regulate neural stem/progenitor cell proliferation by targeting Musashi1
Abstract
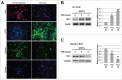
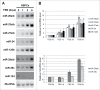
Musashi1 is an RNA binding protein that controls the neural cell fate, being involved in maintaining neural progenitors in their proliferative state. In particular, its downregulation is needed for triggering early neural differentiation programs. In this study, we profiled microRNA expression during the transition from neural progenitors to differentiated astrocytes and underscored 2 upregulated microRNAs, miR-23a and miR-125b, that sinergically act to restrain Musashi1 expression, thus creating a regulatory module controlling neural progenitor proliferation.
Keywords: 3′UTR, 3′Untranslated Region; BrdU, 5-bromo-2′-deoxyuridine; Msi1, Musashi1; NSPCs, Neural Stem/Progenitor Cells; astrocyte differentiation; cell proliferation; miRNA, microRNA; microRNAs; musashi1; neural progenitors; post-transcriptional gene regulation.
Figures


References
-
- Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. J Cell Sci 2002; 115:1355-9; PMID:11896183 - PubMed
-
- Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein musashi-1 in stem cells. Exp Cell Res 2005; 306:349-56; PMID:15925591; http://dx.doi.org/ 10.1016/j.yexcr.2005.02.021 - DOI - PubMed
-
- Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama Y, Miyata T, Okano H. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev Neurosci 2000; 22:139-53; PMID:10657706; http://dx.doi.org/ 10.1159/000017435 - DOI - PubMed
-
- Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H. RNA-binding protein musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci U S A 2002; 99:15194-9; PMID:12407178; http://dx.doi.org/ 10.1073/pnas.232087499 - DOI - PMC - PubMed
-
- Siddall NA, McLaughlin EA, Marriner NL, Hime GR. The RNA-binding protein musashi is required intrinsically to maintain stem cell identity. Proc Natl Acad Sci U S A 2006; 103:8402-7; PMID:16717192; http://dx.doi.org/ 10.1073/pnas.0600906103 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
