Cdt2-mediated XPG degradation promotes gap-filling DNA synthesis in nucleotide excision repair
- PMID: 25483071
- PMCID: PMC4614405
- DOI: 10.4161/15384101.2014.973740
Cdt2-mediated XPG degradation promotes gap-filling DNA synthesis in nucleotide excision repair
Abstract
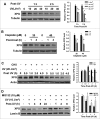
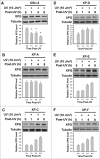
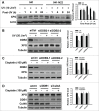
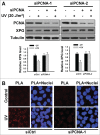
Xeroderma pigmentosum group G (XPG) protein is a structure-specific repair endonuclease, which cleaves DNA strands on the 3' side of the DNA damage during nucleotide excision repair (NER). XPG also plays a crucial role in initiating DNA repair synthesis through recruitment of PCNA to the repair sites. However, the fate of XPG protein subsequent to the excision of DNA damage has remained unresolved. Here, we show that XPG, following its action on bulky lesions resulting from exposures to UV irradiation and cisplatin, is subjected to proteasome-mediated proteolytic degradation. Productive NER processing is required for XPG degradation as both UV and cisplatin treatment-induced XPG degradation is compromised in NER-deficient XP-A, XP-B, XP-C, and XP-F cells. In addition, the NER-related XPG degradation requires Cdt2, a component of an E3 ubiquitin ligase, CRL4(Cdt2). Micropore local UV irradiation and in situ Proximity Ligation assays demonstrated that Cdt2 is recruited to the UV-damage sites and interacts with XPG in the presence of PCNA. Importantly, Cdt2-mediated XPG degradation is crucial to the subsequent recruitment of DNA polymerase δ and DNA repair synthesis. Collectively, our data support the idea of PCNA recruitment to damage sites which occurs in conjunction with XPG, recognition of the PCNA-bound XPG by CRL4(Cdt2) for specific ubiquitylation and finally the protein degradation. In essence, XPG elimination from DNA damage sites clears the chromatin space needed for the subsequent recruitment of DNA polymerase δ to the damage site and completion of gap-filling DNA synthesis during the final stage of NER.
Keywords: CRL4; Cdt2; PCNA; XPG; gap-filling DNA synthesis; nucleotide excision repair; protein degradation; ubiquitylation.
Figures

References
-
- Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev 2006; 106:253-76; PMID:16464005; http://dx.doi.org/ 10.1021/cr040483f - DOI - PubMed
-
- Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks. Trends Genet 2012; 28:566-73; PMID:22824526; http://dx.doi.org/ 10.1016/j.tig.2012.06.004 - DOI - PubMed
-
- Mu D, Park C-H, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem 1995; 270:2415-8; PMID:7852297; http://dx.doi.org/ 10.1074/jbc.270.6.2415 - DOI - PubMed
-
- Araujo SJ, Tirode F, Coin F, Pospiech H, Syvaoja JE, Stucki M, Hubscher U, Egly JM, Wood RD. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev 2000; 14:349-59; PMID:10673506 - PMC - PubMed
-
- Volker M, Mone MJ, Karmakar P, Van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, Van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell 2001; 8:213-24; PMID:11511374; http://dx.doi.org/ 10.1016/S1097-2765(01)00281-7 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
