Quantitative bacterial transcriptomics with RNA-seq
- PMID: 25483350
- PMCID: PMC4323862
- DOI: 10.1016/j.mib.2014.11.011
Quantitative bacterial transcriptomics with RNA-seq
Abstract
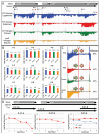
RNA sequencing has emerged as the premier approach to study bacterial transcriptomes. While the earliest published studies analyzed the data qualitatively, the data are readily digitized and lend themselves to quantitative analysis. High-resolution RNA sequence (RNA-seq) data allows transcriptional features (promoters, terminators, operons, among others) to be pinpointed on any bacterial transcriptome. Once the transcriptome is mapped, the activity of transcriptional features can be quantified. Here we highlight how quantitative transcriptome analysis can reveal biological insights and briefly discuss some of the challenges to be faced by the field of bacterial transcriptomics in the near future.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
References
-
- Sharma CM, Vogel J. Differential RNA-seq: the approach behind and the biological insight gained. Curr Opin Microbiol. 2014;19:97–105. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

