Lateral gene transfers and the origins of the eukaryote proteome: a view from microbial parasites
- PMID: 25483352
- PMCID: PMC4728198
- DOI: 10.1016/j.mib.2014.11.018
Lateral gene transfers and the origins of the eukaryote proteome: a view from microbial parasites
Abstract
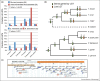
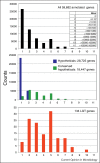
Our knowledge of the extent and functional impact of lateral gene transfer (LGT) from prokaryotes to eukaryotes, outside of endosymbiosis, is still rather limited. Here we review the recent literature, focusing mainly on microbial parasites, indicating that LGT from diverse prokaryotes has played a significant role in the evolution of a number of lineages, and by extension throughout eukaryotic evolution. As might be expected, taxonomic biases for donor prokaryotes indicate that shared habitat is a major factor driving transfers. The LGTs identified predominantly affect enzymes from metabolic pathways, but over a third of LGT are genes for putative proteins of unknown function. Finally, we discuss the difficulties in analysing LGT among eukaryotes and suggest that high-throughput methodologies integrating different approaches are needed to achieve a more global understanding of the importance of LGT in eukaryotic evolution.
Copyright © 2014 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
Similar articles
-
Lateral gene transfer between prokaryotes and eukaryotes.Exp Cell Res. 2017 Sep 15;358(2):421-426. doi: 10.1016/j.yexcr.2017.02.009. Epub 2017 Feb 9. Exp Cell Res. 2017. PMID: 28189637 Free PMC article. Review.
-
Too Much Eukaryote LGT.Bioessays. 2017 Dec;39(12). doi: 10.1002/bies.201700115. Epub 2017 Oct 25. Bioessays. 2017. PMID: 29068466 Review.
-
Old genes in new places: A taxon-rich analysis of interdomain lateral gene transfer events.PLoS Genet. 2022 Jun 22;18(6):e1010239. doi: 10.1371/journal.pgen.1010239. eCollection 2022 Jun. PLoS Genet. 2022. PMID: 35731825 Free PMC article.
-
A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70 % rule.BMC Biol. 2016 Oct 17;14(1):89. doi: 10.1186/s12915-016-0315-9. BMC Biol. 2016. PMID: 27751184 Free PMC article.
-
Grafting or pruning in the animal tree: lateral gene transfer and gene loss?BMC Genomics. 2018 Jun 18;19(1):470. doi: 10.1186/s12864-018-4832-5. BMC Genomics. 2018. PMID: 29914363 Free PMC article.
Cited by
-
Identification of internalin-A-like virulent proteins in Leishmania donovani.Parasit Vectors. 2016 Oct 21;9(1):557. doi: 10.1186/s13071-016-1842-5. Parasit Vectors. 2016. PMID: 27765050 Free PMC article.
-
Identification of constraints influencing the bacterial genomes evolution in the PVC super-phylum.BMC Evol Biol. 2017 Mar 9;17(1):75. doi: 10.1186/s12862-017-0921-3. BMC Evol Biol. 2017. PMID: 28274202 Free PMC article.
-
The role of horizontal gene transfer in the evolution of the oomycetes.PLoS Pathog. 2015 May 28;11(5):e1004805. doi: 10.1371/journal.ppat.1004805. eCollection 2015 May. PLoS Pathog. 2015. PMID: 26020232 Free PMC article. No abstract available.
-
Identification of Key Genes during Ca2+-Induced Genetic Transformation in Escherichia coli by Combining Multi-Omics and Gene Knockout Techniques.Appl Environ Microbiol. 2022 Nov 8;88(21):e0058722. doi: 10.1128/aem.00587-22. Epub 2022 Oct 18. Appl Environ Microbiol. 2022. PMID: 36255244 Free PMC article.
-
Alternative characterizations of Fitch's xenology relation.J Math Biol. 2019 Aug;79(3):969-986. doi: 10.1007/s00285-019-01384-x. Epub 2019 May 20. J Math Biol. 2019. PMID: 31111195
References
-
- Skippington E., Ragan M.A. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol Rev. 2011;35:707–735. - PubMed
-
- Zhaxybayeva O., Doolittle W.F. Lateral gene transfer. Curr Biol. 2011;21:R242–R246. - PubMed
-
- Andersson J.O. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol. 2009;63:177–193. - PubMed
-
- Keeling P.J. Functional and ecological impacts of horizontal gene transfer in eukaryotes. Curr Opin Genet Dev. 2009;19:613–619. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

