Pervasive transcription: detecting functional RNAs in bacteria
- PMID: 25483405
- PMCID: PMC4581347
- DOI: 10.4161/21541272.2014.944039
Pervasive transcription: detecting functional RNAs in bacteria
Abstract
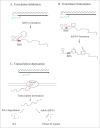
Pervasive, or genome-wide, transcription has been reported in all domains of life. In bacteria, most pervasive transcription occurs antisense to protein-coding transcripts, although recently a new class of pervasive RNAs was identified that originates from within annotated genes. Initially considered to be non-functional transcriptional noise, pervasive transcription is increasingly being recognized as important in regulating gene expression. The function of pervasive transcription is an extensively debated question in the field of transcriptomics and regulatory RNA biology. Here, we highlight the most recent contributions addressing the purpose of pervasive transcription in bacteria and discuss their implications.
Keywords: Hfq; RNA; RNase III; antisense transcription; asRNA; asRNA, antisense RNA; double-stranded RNA; dsRNA, double-stranded RNA; intraRNA, intragenic RNA; ncRNA, non-coding RNA; pervasive transcription; promoter; sigma factor; spurious transcription; transcriptome.
Figures
References
-
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. . Landscape of transcription in human cells. Nature 2012; 489:101-8; PMID:22955620; http://dx.doi.org/10.1038/nature11233 - DOI - PMC - PubMed
-
- Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 2011; 75:286-300; PMID:21646430; http://dx.doi.org/10.1128/MMBR.00032-10 - DOI - PMC - PubMed
-
- Wagner EG, Simons RW. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol 1994; 48:713-42; PMID:7826024; http://dx.doi.org/10.1146/annurev.mi.48.100194.003433 - DOI - PubMed
-
- Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet 2010; 44:167-88; PMID:20707673; http://dx.doi.org/10.1146/annurev-genet-102209-163523 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
