Autophagy-dependent PELI3 degradation inhibits proinflammatory IL1B expression
- PMID: 25483963
- PMCID: PMC4502728
- DOI: 10.4161/auto.32178
Autophagy-dependent PELI3 degradation inhibits proinflammatory IL1B expression
Abstract
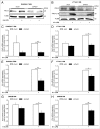
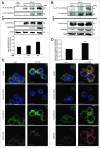
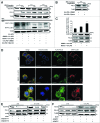
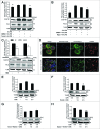
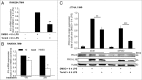
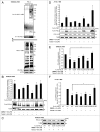
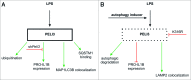
Lipopolysaccharide (LPS)-induced activation of TLR4 (toll-like receptor 4) is followed by a subsequent overwhelming inflammatory response, a hallmark of the first phase of sepsis. Therefore, counteracting excessive innate immunity by autophagy is important to contribute to the termination of inflammation. However, the exact molecular details of this interplay are only poorly understood. Here, we show that PELI3/Pellino3 (pellino E3 ubiquitin protein ligase family member 3), which is an E3 ubiquitin ligase and scaffold protein in TLR4-signaling, is impacted by autophagy in macrophages (MΦ) after LPS stimulation. We noticed an attenuated mRNA expression of proinflammatory Il1b (interleukin 1, β) in Peli3 knockdown murine MΦ in response to LPS treatment. The autophagy adaptor protein SQSTM1/p62 (sequestosome 1) emerged as a potential PELI3 binding partner in TLR4-signaling. siRNA targeting Sqstm1 and Atg7 (autophagy related 7), pharmacological inhibition of autophagy by wortmannin as well as blocking the lysosomal vacuolar-type H(+)-ATPase by bafilomycin A1 augmented PELI3 protein levels, while inhibition of the proteasome had no effect. Consistently, treatment to induce autophagy by MTOR (mechanistic target of rapamycin (serine/threonine kinase)) inhibition or starvation enhanced PELI3 degradation and reduced proinflammatory Il1b expression. PELI3 was found to be ubiquitinated upon LPS stimulation and point mutation of PELI3-lysine residue 316 (Lys316Arg) attenuated Torin2-dependent degradation of PELI3. Immunofluorescence analysis revealed that PELI3 colocalized with the typical autophagy markers MAP1LC3B/LC3B (microtubule-associated protein 1 light chain 3 β) and LAMP2 (lysosomal-associated membrane protein 2). Our observations suggest that autophagy causes PELI3 degradation during TLR4-signaling, thereby impairing the hyperinflammatory phase during sepsis.
Keywords: ACTB, actin, beta; ATG7, autophagy-related 7; BECN1, Beclin 1, autophagy related; Baf A1, bafilomycin A1; CHX, cycloheximide; Epoxo, epoxomycin; HBSS, Hank's balanced salt solution; IL1B, interleukin 1, beta; IRAKs, interleukin-1 receptor-associated kinases; LAMP2, lysosomal-associated membrane protein 2; LPS; LPS, lipopolysaccharide; MAP1LC3B, microtubule-associated protein 1 light chain 3 beta; MAP3K14, mitogen-activated protein kinase kinase kinase 14; MAP3K7, mitogen-activated protein kinase kinase kinase 7; MAPK1/3, mitogen-activated protein kinase 1/3; MAPK14, mitogen-activated protein kinase 14; MAPK8/9, mitogen-activated protein kinase 8/9; MTOR, mechanistic target of rapamycin (serine/threonine kinase); MYD88, myeloid differentiation primary response 88; MΦ, macrophages; NFE2L2, nuclear factor, erythroid 2-like 2; NFKB, nuclear factor of kappa light polypeptide gene enhancer in B-cells; NLRP3, NLR family, pyrin domain containing 3; PELI3; PELI3, pellino E3 ubiquitin protein ligase family member 3; PRRs, pattern recognition receptors; RIPK1, receptor (TNFRSF)-interacting serine-threonine kinase 1; Rapa, rapamycin; SQSTM1; SQSTM1, sequestosome 1; TABs, TGF-beta activated kinase 1/MAP3K7 binding protein 1/2/3; TBK1, TANK-binding kinase 1; TICAM1, toll-like receptor adaptor molecule 1; TLRs, toll-like receptors; TNF, tumor necrosis factor; TRAF6, TNF receptor-associated factor 6, E3 ubiquitin protein ligase; TUBB, tubulin, beta class I; Torin2; UBB, ubiquitin B; WT, wildtype; Wortm, wortmannin; autophagy.
Figures
Similar articles
-
The GST-BHMT assay reveals a distinct mechanism underlying proteasome inhibition-induced macroautophagy in mammalian cells.Autophagy. 2015;11(5):812-32. doi: 10.1080/15548627.2015.1034402. Autophagy. 2015. PMID: 25984893 Free PMC article.
-
Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination.Autophagy. 2014;10(12):2239-50. doi: 10.4161/15548627.2014.981792. Autophagy. 2014. PMID: 25484083 Free PMC article.
-
Inactivation of MTOR promotes autophagy-mediated epithelial injury in particulate matter-induced airway inflammation.Autophagy. 2020 Mar;16(3):435-450. doi: 10.1080/15548627.2019.1628536. Epub 2019 Jun 16. Autophagy. 2020. PMID: 31203721 Free PMC article.
-
Organelle-specific autophagy in inflammatory diseases: a potential therapeutic target underlying the quality control of multiple organelles.Autophagy. 2021 Feb;17(2):385-401. doi: 10.1080/15548627.2020.1725377. Epub 2020 Feb 12. Autophagy. 2021. PMID: 32048886 Free PMC article. Review.
-
The exploitation of host autophagy and ubiquitin machinery by Mycobacterium tuberculosis in shaping immune responses and host defense during infection.Autophagy. 2023 Jan;19(1):3-23. doi: 10.1080/15548627.2021.2021495. Epub 2022 Jan 9. Autophagy. 2023. PMID: 35000542 Free PMC article. Review.
Cited by
-
LL-37 Exacerbates Local Inflammation in Sepsis-Induced Acute Lung Injury by Preventing Mitochondrial DNA (mtDNA) Degradation-Induced Autophagy.Med Sci Monit. 2019 Aug 18;25:6193-9203. doi: 10.12659/MSM.915298. Med Sci Monit. 2019. PMID: 31422413 Free PMC article.
-
PD-L1 maintains neutrophil extracellular traps release by inhibiting neutrophil autophagy in endotoxin-induced lung injury.Front Immunol. 2022 Aug 9;13:949217. doi: 10.3389/fimmu.2022.949217. eCollection 2022. Front Immunol. 2022. PMID: 36016930 Free PMC article.
-
Autophagy down regulates pro-inflammatory mediators in BV2 microglial cells and rescues both LPS and alpha-synuclein induced neuronal cell death.Sci Rep. 2017 Mar 3;7:43153. doi: 10.1038/srep43153. Sci Rep. 2017. PMID: 28256519 Free PMC article.
-
Emerging Roles of Disabled Homolog 2 (DAB2) in Immune Regulation.Front Immunol. 2020 Oct 15;11:580302. doi: 10.3389/fimmu.2020.580302. eCollection 2020. Front Immunol. 2020. PMID: 33178208 Free PMC article. Review.
-
Hydrogen alleviates mitochondrial dysfunction and organ damage via autophagy‑mediated NLRP3 inflammasome inactivation in sepsis.Int J Mol Med. 2019 Oct;44(4):1309-1324. doi: 10.3892/ijmm.2019.4311. Epub 2019 Aug 13. Int J Mol Med. 2019. PMID: 31432098 Free PMC article.
References
-
- Medzhitov R, Janeway C, Jr. Innate immunity. N Engl J Med 2000; 343:338-44; PMID:10922424; http://dx.doi.org/10.1056/NEJM200008033430506 - DOI - PubMed
-
- Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 2009; 22:240-73; PMID:19366914; http://dx.doi.org/10.1128/CMR.00046-08 - DOI - PMC - PubMed
-
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol 2004; 4:499-511; PMID:15229469; http://dx.doi.org/10.1038/nri1391 - DOI - PubMed
-
- Pandey S, Agrawal DK. Immunobiology of Toll-like receptors: emerging trends. Immunol Cell Biol 2006; 84:333-41; PMID:16834572; http://dx.doi.org/10.1111/j.1440-1711.2006.01444.x - DOI - PubMed
-
- O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 2007; 7:353-64; PMID:17457343; http://dx.doi.org/10.1038/nri2079 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
