Genome-wide DNA methylation analysis in precursor B-cells
- PMID: 25484143
- PMCID: PMC4622941
- DOI: 10.4161/15592294.2014.983379
Genome-wide DNA methylation analysis in precursor B-cells
Abstract
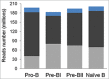
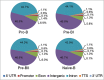
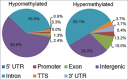
DNA methylation is responsible for regulating gene expression and cellular differentiation and for maintaining genomic stability during normal human development. Furthermore, it plays a significant role in the regulation of hematopoiesis. In order to elucidate the influence of DNA methylation during B-cell development, genome-wide DNA methylation status of pro-B, pre-BI, pre-BII, and naïve-B-cells isolated from human umbilical cord blood was determined using the methylated CpG island recovery assay followed by next generation sequencing. On average, 182-200 million sequences were generated for each precursor B-cell subset in 10 biological replicates. An overall decrease in methylation was observed during the transition from pro-B to pre-BI, whereas no differential methylation was observed in the pre-BI to pre-BII transition or in the pre-BII to naïve B-cell transition. Most of the methylated regions were located within intergenic and intronic regions not present in a CpG island context. Putative novel enhancers were identified in these regions that were differentially methylated between pro-B and pre-BI cells. The genome-wide methylation profiles are publically available and may be used to gain a better understanding of the involvement of atypical DNA methylation in the pathogenesis of malignancies associated with precursor B-cells.
Keywords: CG dinucleotide; CLP, common lymphoid progenitor cells; CpGI, CpG island; DMRs, differentially methylated regions; DNA methylation; FDR, false discovery rate.; H3K27ac, histone H3 lysine 27 acetylation; H3K4me1, histone H3 lysine 4 monomethylation; HCB, human umbilical cord blood; HSCs, haematopoietic stem cells; MBDs, methyl CpG binding domains; MIRA-seq, methylated CpG island recovery assay (MIRA) followed by next generation sequencing; MeCP2, methyl CpG binding protein 2; Pre-B, precursor B-cell; CD; Pro-B, progenitor B-cell; ROIs, regions of interest; TFs, transcription factors; acute lymphoblastic leukemia; CpG; cluster of differentiation; ALL; enhancer; next-generation sequencing; precursor B-cell; umbilical cord blood.
Figures


References
-
- Hystad ME, Myklebust JH, Bø TH, Sivertsen EA, Rian E, Forfang L, Munthe E, Rosenwald A, Chiorazzi M, Jonassen I, et al. . Characterization of early stages of human B cell development by gene expression profiling. J Immunol 2007; 179(6):3662-71; PMID:17785802; http://dx.doi.org/10.4049/jimmunol.179.6.3662. - DOI - PubMed
-
- van Zelm MC, van der Burg M, de Ridder D, Barendregt BH, de Haas EF, Reinders MJ, Lankester AC, Révész T, Staal FJ, van Dongen JJ. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol 2005; 175(9):5912-22; PMID:16237084; http://dx.doi.org/10.4049/jimmunol.175.9.5912. - DOI - PubMed
-
- Pérez-Vera P, Reyes-León A, Fuentes-Pananá EM. Signaling proteins and transcription factors in normal and malignant early B cell development. Bone Marrow Res 2011; 2011:502751; http://dx.doi.org/10.1155/2011/502751. - DOI - PMC - PubMed
-
- Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat Rev Immunol 2005; 5(6):497-508; PMID:15928681; http://dx.doi.org/10.1038/nri1633. - DOI - PubMed
-
- Harmston N, Lenhard B. Chromatin and epigenetic features of long-range gene regulation. Nucleic Acids Res 2013; 41(15):7185-99; PMID:23766291; http://dx.doi.org/10.1093/nar/gkt499. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
