Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production
- PMID: 25484880
- PMCID: PMC4240174
- DOI: 10.3389/fmicb.2014.00636
Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production
Abstract
Bacillus amyloliquefaciens SQR9 exhibited predominantly antagonistic activities against a broad range of soilborne pathogens. The fungi-induced SQR9 extracts possess stronger antifungal activities compared with SQR9 monoculture extracts. To investigate how SQR9 fine-tunes lipopeptides (LPs) and a siderophore bacillibactin production to control different fungal pathogens, LPs and bacillibactin production and transcription of the respective encoding genes in SQR9 were measured and compared with six different soilborne fungal pathogens. SQR9 altered its spectrum of antifungal compounds production responding to different fungal pathogen. Bacillomycin D was the major LP produced when SQR9 was confronted with Fusarium oxysporum. Fengycin contributed to the antagonistic activity against Verticillium dahliae kleb, Fusarium oxysporum, Fusarium solani, and Phytophthora parasitica. Surfactin participated in the antagonistic process against Sclerotinia sclerotiorum, Rhizoctonia solani, and Fusarium solani. Bacillibactin was up-regulated when SQR9 was confronted with all tested fungi. The reduction in antagonistic activities of three LP and bacillibactin deficient mutants of SQR9 when confronted with the six soilborne fungal pathogens provided further evidence of the contribution of LPs and bacillibactin in controlling fungal pathogens. These results provide a new understanding of specific cues in bacteria-fungi interactions and provide insights for agricultural applications.
Keywords: Bacillus amyloliquefaciens SQR9; bacteria-fungal interaction; lipopeptide antibiotics; soilborne pathogens; transcriptional response.
Figures
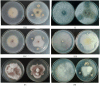

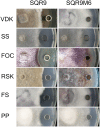
References
-
- Barret M., Frey-Klett P., Guillerm-Erckelboudt A. Y., Boutin M., Guernec G., Sarniguet A. (2009). Effect of wheat roots infected with the pathogenic fungus Gaeumannomyces graminis var. tritici on gene expression of the biocontrol bacterium Pseudomonas fluorescens Pf29Arp. Mol. Plant Microbe Interact. 22, 1611–1623. 10.1094/MPMI-22-12-1611 - DOI - PubMed
-
- Becker D. M., Kinkel L. L., Schottel J. L. (1997). Evidence for interspecies communication and its potential role in pathogen suppression in a naturally occurring disease suppressive soil. Can. J. Microbiol. 43, 985–990 10.1139/m97-142 - DOI
-
- Behnam S., Ahmadzadeh M., Sharifi Tehrani A., Hedjaroude G. A., Farzaneh M. (2006). Biological control of Sclerotinia sclerotiorum (Lib.) de Bary, the causal agent of white mold, by Pseudomonas species on canola petals. Commun. Agric. Appl. Biol. Sci. 72, 993–996. 10.1016/j.cropro.2006.04.007 - DOI - PubMed
-
- Cao Y., Zhang Z., Ling N., Yuan Y., Zheng X., Shen B., et al. (2011). Bacillus subtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol. Fertil. Soils 47, 495–506 10.1007/s00374-011-0556-2 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

