Genomic prediction in an admixed population of Atlantic salmon (Salmo salar)
- PMID: 25484890
- PMCID: PMC4240172
- DOI: 10.3389/fgene.2014.00402
Genomic prediction in an admixed population of Atlantic salmon (Salmo salar)
Abstract
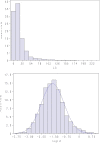
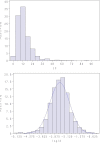
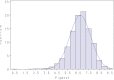
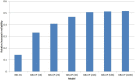
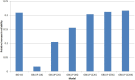
Reliability of genomic selection (GS) models was tested in an admixed population of Atlantic salmon, originating from crossing of several wild subpopulations. The models included ordinary genomic BLUP models (GBLUP), using genome-wide SNP markers of varying densities (1-220 k), a genomic identity-by-descent model (IBD-GS), using linkage analysis of sparse genome-wide markers, as well as a classical pedigree-based model. Reliabilities of the models were compared through 5-fold cross-validation. The traits studied were salmon lice (Lepeophtheirus salmonis) resistance (LR), measured as (log) density on the skin and fillet color (FC), with respective estimated heritabilities of 0.14 and 0.43. All genomic models outperformed the classical pedigree-based model, for both traits and at all marker densities. However, the relative improvement differed considerably between traits, models and marker densities. For the highly heritable FC, the IBD-GS had similar reliability as GBLUP at high marker densities (>22 k). In contrast, for the lowly heritable LR, IBD-GS was clearly inferior to GBLUP, irrespective of marker density. Hence, GBLUP was robust to marker density for the lowly heritable LR, but sensitive to marker density for the highly heritable FC. We hypothesize that this phenomenon may be explained by historical admixture of different founder populations, expected to reduce short-range lice density (LD) and induce long-range LD. The relative importance of LD/relationship information is expected to decrease/increase with increasing heritability of the trait. Still, using the ordinary GBLUP, the typical long-range LD of an admixed population may be effectively captured by sparse markers, while efficient utilization of relationship information may require denser markers (e.g., 22 k or more).
Keywords: Atlantic salmon; admixture; genetics; genomic selection; reliability.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

