Genetics of Capsular Polysaccharides and Cell Envelope (Glyco)lipids
- PMID: 25485178
- PMCID: PMC4255156
- DOI: 10.1128/microbiolspec.MGM2-0021-2013
Genetics of Capsular Polysaccharides and Cell Envelope (Glyco)lipids
Abstract
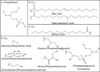
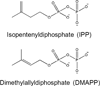
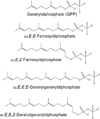
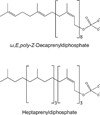
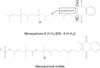
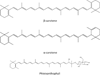
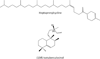
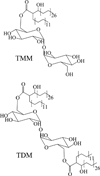
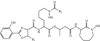
This chapter summarizes what is currently known of the structures, physiological roles, involvement in pathogenicity and biogenesis of a variety of non-covalently bound cell envelope lipids and glycoconjugates of Mycobacterium tuberculosis and other Mycobacterium species. Topics addressed in this chapter include phospholipids; phosphatidylinositol mannosides; triglycerides; isoprenoids and related compounds (polyprenyl phosphate, menaquinones, carotenoids, non-carotenoid cyclic isoprenoids); acyltrehaloses (lipooligosaccharides, trehalose mono- and di-mycolates, sulfolipids, di- and poly-acyltrehaloses); mannosyl-beta-1-phosphomycoketides; glycopeptidolipids; phthiocerol dimycocerosates, para-hydroxybenzoic acids and phenolic glycolipids; mycobactins; mycolactones; and capsular polysaccharides.
Figures






Similar articles
-
Genetics of Capsular Polysaccharides and Cell Envelope (Glyco)lipids.Microbiol Spectr. 2014 Aug;2(4):MGM2-0021-2013. doi: 10.1128/microbiolspec.MGM2-0021-2013. Microbiol Spectr. 2014. PMID: 26104202 Review.
-
Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species.J Bacteriol. 1996 Jan;178(2):456-61. doi: 10.1128/jb.178.2.456-461.1996. J Bacteriol. 1996. PMID: 8550466 Free PMC article.
-
Molecular dissection of the biosynthetic relationship between phthiocerol and phthiodiolone dimycocerosates and their critical role in the virulence and permeability of Mycobacterium tuberculosis.FEBS J. 2007 Apr;274(8):1957-69. doi: 10.1111/j.1742-4658.2007.05740.x. Epub 2007 Mar 19. FEBS J. 2007. PMID: 17371506
-
Analysis of the Lipid Composition of Mycobacteria by Thin Layer Chromatography.J Vis Exp. 2021 Apr 16;(170). doi: 10.3791/62368. J Vis Exp. 2021. PMID: 33938877
-
The envelope of mycobacteria.Annu Rev Biochem. 1995;64:29-63. doi: 10.1146/annurev.bi.64.070195.000333. Annu Rev Biochem. 1995. PMID: 7574484 Review.
Cited by
-
Transporters Involved in the Biogenesis and Functionalization of the Mycobacterial Cell Envelope.Chem Rev. 2021 May 12;121(9):5124-5157. doi: 10.1021/acs.chemrev.0c00869. Epub 2020 Nov 10. Chem Rev. 2021. PMID: 33170669 Free PMC article. Review.
-
Understanding Metabolic Regulation Between Host and Pathogens: New Opportunities for the Development of Improved Therapeutic Strategies Against Mycobacterium tuberculosis Infection.Front Cell Infect Microbiol. 2021 Mar 16;11:635335. doi: 10.3389/fcimb.2021.635335. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33796480 Free PMC article. Review.
-
Trehalose polyphleates participate in Mycobacterium abscessus fitness and pathogenesis.mBio. 2024 Dec 11;15(12):e0297024. doi: 10.1128/mbio.02970-24. Epub 2024 Oct 30. mBio. 2024. PMID: 39475242 Free PMC article.
-
Functional Amyloid and Other Protein Fibers in the Biofilm Matrix.J Mol Biol. 2018 Oct 12;430(20):3642-3656. doi: 10.1016/j.jmb.2018.07.026. Epub 2018 Aug 8. J Mol Biol. 2018. PMID: 30098341 Free PMC article. Review.
-
The Role of Phosphatidylinositol Mannosides in the Serological Diagnosis of Mycobacterial Infections.Vet Sci. 2019 Nov 13;6(4):91. doi: 10.3390/vetsci6040091. Vet Sci. 2019. PMID: 31766256 Free PMC article.
References
-
- Daffé M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol. 1998;39:131–203. - PubMed
-
- Sani M, Houben ENG, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jimenez CR, Daffe M, Appelmelk BJ, Bitter W, van der Wel N, Peters PJ. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010;6:e1000794. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

