RFTS-deleted DNMT1 enhances tumorigenicity with focal hypermethylation and global hypomethylation
- PMID: 25485502
- PMCID: PMC4615144
- DOI: 10.4161/15384101.2014.950886
RFTS-deleted DNMT1 enhances tumorigenicity with focal hypermethylation and global hypomethylation
Abstract
Site-specific hypermethylation of tumor suppressor genes accompanied by genome-wide hypomethylation are epigenetic hallmarks of malignancy. However, the molecular mechanisms that drive these linked changes in DNA methylation remain obscure. DNA methyltransferase 1 (DNMT1), the principle enzyme responsible for maintaining methylation patterns is commonly dysregulated in tumors. Replication foci targeting sequence (RFTS) is an N-terminal domain of DNMT1 that inhibits DNA-binding and catalytic activity, suggesting that RFTS deletion would result in a gain of DNMT1 function. However, a substantial body of data suggested that RFTS is required for DNMT1 activity. Here, we demonstrate that deletion of RFTS alters DNMT1-dependent DNA methylation during malignant transformation. Compared to full-length DNMT1, ectopic expression of hyperactive DNMT1-ΔRFTS caused greater malignant transformation and enhanced promoter methylation with condensed chromatin structure that silenced DAPK and DUOX1 expression. Simultaneously, deletion of RFTS impaired DNMT1 chromatin association with pericentromeric Satellite 2 (SAT2) repeat sequences and produced DNA demethylation at SAT2 repeats and globally. To our knowledge, RFTS-deleted DNMT1 is the first single factor that can reprogram focal hypermethylation and global hypomethylation in parallel during malignant transformation. Our evidence suggests that the RFTS domain of DNMT1 is a target responsible for epigenetic changes in cancer.
Keywords: COSMIC, catalog of somatic mutation in cancer; DNA methylation; DNMT1; DNMT1, DNA methyltransferase 1; HELP, HpaII tiny fragment enrichment by ligation-mediated PCR; KEGG, Kyoto Encyclopedia of Genes and Genomes; RFTS domain; RFTS, replication foci targeting sequence; TSGs, tumor suppressor genes; chromatin; tumorigenicity.
Figures

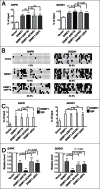

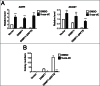
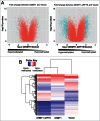


Comment in
-
DNMT1-associated DNA methylation changes in cancer.Cell Cycle. 2015;14(1):5. doi: 10.4161/15384101.2014.989963. Cell Cycle. 2015. PMID: 25483051 Free PMC article. No abstract available.
References
-
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005; 6:597-610; PMID:16136652; http://dx.doi.org/ 10.1038/nrg1655 - DOI - PubMed
-
- Issa J-P. CpG island methylator phenotype in cancer. Nat Rev Cancer 2004; 4:988-93; PMID:15573120 - PubMed
-
- Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer 2004; 4:707-17; PMID:15343277; http://dx.doi.org/ 10.1038/nrc1432 - DOI - PubMed
-
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3:415-28; PMID:12042769 - PubMed
-
- Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res 1988; 48:1159-61; PMID:3342396 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
