Temporal separation of replication and transcription during S-phase progression
- PMID: 25485504
- PMCID: PMC4615114
- DOI: 10.4161/15384101.2014.953876
Temporal separation of replication and transcription during S-phase progression
Abstract
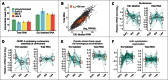
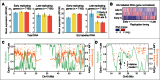
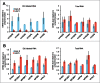
Transcriptional events during S-phase are critical for cell cycle progression. Here, by using a nascent RNA capture assay coupled with high-throughput sequencing, we determined the temporal patterns of transcriptional events that occur during S-phase. We show that genes involved in critical S-phase-specific biological processes such as nucleosome assembly and DNA repair have temporal transcription patterns across S-phase that are not evident from total RNA levels. By comparing transcription timing with replication timing in S-phase, we show that early replicating genes show increased transcription late in S-phase whereas late replicating genes are predominantly transcribed early in S-phase. Global anti-correlation between replication and transcription timing was observed only based on nascent RNA but not total RNA. Our data provides a detailed view of ongoing transcriptional events during the S-phase of cell cycle, and supports that transcription and replication are temporally separated.
Keywords: Cell cycle; Ethynyl Uridine, EtU; Replication; Replication timing; S-phase; Transcription.
Figures

References
-
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol biol Cell 2002; 13:1977-2000; PMID:12058064; http://dx.doi.org/ 10.1091/mbc.02-02-0030 - DOI - PMC - PubMed
-
- Grant GD, Brooks L, 3rd, Zhang X, Mahoney JM, Martyanov V, Wood TA, Sherlock G, Cheng C, Whitfield ML. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol biol Cell 2013; 24:3634-50; PMID:24109597; http://dx.doi.org/ 10.1091/mbc.E13-05-0264 - DOI - PMC - PubMed
-
- Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge SJ, Davis RW, Lockhart DJ. Transcriptional regulation and function during the human cell cycle. Nat Genet 2001; 27:48-54; PMID:11137997 - PubMed
-
- van der Meijden CM, Lapointe DS, Luong MX, Peric-Hupkes D, Cho B, Stein JL, van Wijnen AJ, Stein GS. Gene profiling of cell cycle progression through S-phase reveals sequential expression of genes required for DNA replication and nucleosome assembly. Cancer Res 2002; 62:3233-43; PMID:12036939 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
