Follicular helper T cell signature in type 1 diabetes
- PMID: 25485678
- PMCID: PMC4382272
- DOI: 10.1172/JCI76238
Follicular helper T cell signature in type 1 diabetes
Abstract
The strong genetic association between particular HLA alleles and type 1 diabetes (T1D) indicates a key role for CD4+ T cells in disease; however, the differentiation state of the responsible T cells is unclear. T cell differentiation originally was considered a dichotomy between Th1 and Th2 cells, with Th1 cells deemed culpable for autoimmune islet destruction. Now, multiple additional T cell differentiation fates are recognized with distinct roles. Here, we used a transgenic mouse model of diabetes to probe the gene expression profile of islet-specific T cells by microarray and identified a clear follicular helper T (Tfh) cell differentiation signature. Introduction of T cells with a Tfh cell phenotype from diabetic animals efficiently transferred diabetes to recipient animals. Furthermore, memory T cells from patients with T1D expressed elevated levels of Tfh cell markers, including CXCR5, ICOS, PDCD1, BCL6, and IL21. Defects in the IL-2 pathway are associated with T1D, and IL-2 inhibits Tfh cell differentiation in mice. Consistent with these previous observations, we found that IL-2 inhibited human Tfh cell differentiation and identified a relationship between IL-2 sensitivity in T cells from patients with T1D and acquisition of a Tfh cell phenotype. Together, these findings identify a Tfh cell signature in autoimmune diabetes and suggest that this population could be used as a biomarker and potentially targeted for T1D interventions.
Figures



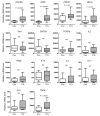



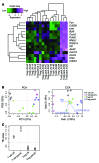
Comment in
-
Immunology: Follicular helper T cells in T1DM.Nat Rev Endocrinol. 2015 Feb;11(2):65. doi: 10.1038/nrendo.2014.216. Epub 2014 Dec 9. Nat Rev Endocrinol. 2015. PMID: 25488481 No abstract available.
References
-
- Rabinovitch A, Suarez-Pinzon WL, Sorensen O, Bleackley RC, Power RF. IFN-gamma gene expression in pancreatic islet-infiltrating mononuclear cells correlates with autoimmune diabetes in nonobese diabetic mice. J Immunol. 1995;154(9):4874–4882. - PubMed
-
- Kallmann BA, et al. Systemic bias of cytokine production toward cell-mediated immune regulation in IDDM and toward humoral immunity in Graves’ disease. Diabetes. 1997;46(2):237–243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

