Identification of the interactors of human nibrin (NBN) and of its 26 kDa and 70 kDa fragments arising from the NBN 657del5 founder mutation
- PMID: 25485873
- PMCID: PMC4259352
- DOI: 10.1371/journal.pone.0114651
Identification of the interactors of human nibrin (NBN) and of its 26 kDa and 70 kDa fragments arising from the NBN 657del5 founder mutation
Abstract
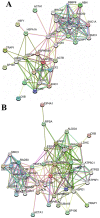
Nibrin (also named NBN or NBS1) is a component of the MRE11/RAD50/NBN complex, which is involved in early steps of DNA double strand breaks sensing and repair. Mutations within the NBN gene are responsible for the Nijmegen breakage syndrome (NBS). The 90% of NBS patients are homozygous for the 657del5 mutation, which determines the synthesis of two truncated proteins of 26 kDa (p26) and 70 kDa (p70). Here, HEK293 cells have been exploited to transiently express either the full-length NBN protein or the p26 or p70 fragments, followed by affinity chromatography enrichment of the eluates. The application of an unsupervised proteomics approach, based upon SDS-PAGE separation and shotgun digestion of protein bands followed by MS/MS protein identification, indicates the occurrence of previously unreported protein interacting partners of the full-length NBN protein and the p26 fragment containing the FHA/BRCT1 domains, especially after cell irradiation. In particular, results obtained shed light on new possible roles of NBN and of the p26 fragment in ROS scavenging, in the DNA damage response, and in protein folding and degradation. In particular, here we show that p26 interacts with PARP1 after irradiation, and this interaction exerts an inhibitory effect on PARP1 activity as measured by NAD+ levels. Furthermore, the p26-PARP1 interaction seems to be responsible for the persistence of ROS, and in turn of DSBs, at 24 h from IR. Since some of the newly identified interactors of the p26 and p70 fragments have not been found to interact with the full-length NBN, these interactions may somehow contribute to the key biological phenomena underpinning NBS.
Conflict of interest statement
Figures





References
-
- D'Amours D, Jackson SP (2002) The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol 3:317–327. - PubMed
-
- Demuth I, Frappart PO, Hildebrand G, Melchers A, Lobitz S, et al. (2004) An inducible null mutant murine model of Nijmegen breakage syndrome proves the essential function of NBS1 in chromosomal stability and cell viability. Hum Mol Genet 13:2385–2397. - PubMed
-
- Digweed M, Sperling K (2004) Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair (Amst) 3:1207–1217. - PubMed
-
- Kobayashi J, Antoccia A, Tauchi H, Matsuura S, Komatsu K (2004) NBS1 and its functional role in the DNA damage response. DNA Repair(Amst) 3:855–861. - PubMed
-
- Iijima K, Ohara M, Seki R, Tauchi H (2008) Dancing on damaged chromatin: functions of ATM and the RAD50/MRE11/NBS1 complex in cellular responses to DNA damage. J Radiat Res 49:451–464. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

