Molecular cloning, characterization and positively selected sites of the glutathione S-transferase family from Locusta migratoria
- PMID: 25486043
- PMCID: PMC4259467
- DOI: 10.1371/journal.pone.0114776
Molecular cloning, characterization and positively selected sites of the glutathione S-transferase family from Locusta migratoria
Abstract
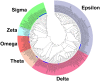
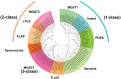
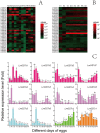
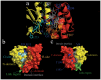
Glutathione S-transferases (GSTs) are multifunctional enzymes that are involved in the metabolism of endogenous and exogenous compounds and are related to insecticide resistance. The purpose of this study was to provide new information on the molecular characteristics and the positive selection of locust GSTs. Based on the transcriptome database, we sequenced 28 cytosolic GSTs and 4 microsomal GSTs from the migratory locust (Locusta migratoria). We assigned the 28 cytosolic GSTs into 6 classes--sigma, epsilon, delta, theta, omega and zeta, and the 4 microsomal GSTs into 2 subclasses--insect and MGST3. The tissue- and stage-expression patterns of the GSTs differed at the mRNA level. Further, the substrate specificities and kinetic constants of the cytosolic GSTs differed markedly at the protein level. The results of likelihood ratio tests provided strong evidence for positive selection in the delta class. The result of Bayes Empirical Bayes analysis identified 4 amino acid sites in the delta class as positive selection sites. These sites were located on the protein surface. Our findings will facilitate the elucidation of the molecular characteristics and evolutionary aspects of insect GST superfamily.
Conflict of interest statement
Figures
Similar articles
-
Effects of chlorpyrifos on glutathione S-transferase in migratory locust, Locusta migratoria.Pestic Biochem Physiol. 2014 Feb;109:1-5. doi: 10.1016/j.pestbp.2013.12.008. Epub 2014 Jan 18. Pestic Biochem Physiol. 2014. PMID: 24581378
-
Identification and characterisation of ten glutathione S-transferase genes from oriental migratory locust, Locusta migratoria manilensis (Meyen).Pest Manag Sci. 2011 Jun;67(6):697-704. doi: 10.1002/ps.2110. Epub 2011 Mar 16. Pest Manag Sci. 2011. PMID: 21413139
-
Heterologous expression and characterization of a sigma glutathione S-transferase involved in carbaryl detoxification from oriental migratory locust, Locusta migratoria manilensis (Meyen).J Insect Physiol. 2012 Feb;58(2):220-7. doi: 10.1016/j.jinsphys.2011.10.011. Epub 2011 Nov 2. J Insect Physiol. 2012. PMID: 22075389
-
Mosquito glutathione transferases.Methods Enzymol. 2005;401:226-41. doi: 10.1016/S0076-6879(05)01014-1. Methods Enzymol. 2005. PMID: 16399389 Review.
-
The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors.Curr Opin Insect Sci. 2018 Jun;27:97-102. doi: 10.1016/j.cois.2018.04.007. Epub 2018 Apr 24. Curr Opin Insect Sci. 2018. PMID: 30025642 Review.
Cited by
-
De Novo Assembly and Characterization of the Transcriptome of Grasshopper Shirakiacris shirakii.Int J Mol Sci. 2016 Jul 22;17(7):1110. doi: 10.3390/ijms17071110. Int J Mol Sci. 2016. PMID: 27455245 Free PMC article.
-
Differential detoxification enzyme profiles in C-corn strain and R-rice strain of Spodoptera frugiperda by comparative genomic analysis: insights into host adaptation.BMC Genomics. 2025 Jan 6;26(1):14. doi: 10.1186/s12864-024-11185-2. BMC Genomics. 2025. PMID: 39762739 Free PMC article.
-
Effects of phenol on glutathione S-transferase expression and enzyme activity in Chironomus kiiensis larvae.Ecotoxicology. 2019 Sep;28(7):754-762. doi: 10.1007/s10646-019-02071-9. Epub 2019 Jun 28. Ecotoxicology. 2019. PMID: 31254185
-
Multiple Glutathione S-Transferase Genes in Heortia vitessoides (Lepidoptera: Crambidae): Identification and Expression Patterns.J Insect Sci. 2018 May 1;18(3):23. doi: 10.1093/jisesa/iey064. J Insect Sci. 2018. PMID: 29912411 Free PMC article.
-
Heortia vitessoides Infests Aquilaria sinensis: A Systematic Review of Climate Drivers, Management Strategies, and Molecular Mechanisms.Insects. 2025 Jul 2;16(7):690. doi: 10.3390/insects16070690. Insects. 2025. PMID: 40725320 Free PMC article. Review.
References
-
- Yang ML, Zhang JZ, Zhu KY, Xuan T, Liu XJ, et al. (2009) Mechanisms of organophosphate resistance in a field population of oriental migratory locust, Locusta migratoria manilensis (Meyen). Archives of Insect Biochemistry and Physiology 71:3–15. - PubMed
-
- Zhang X, Li T, Zhang J, Li D, Guo Y, et al. (2012) Structural and catalytic role of two conserved tyrosines in delta-class glutathione S-transferase from Locusta migratoria . Archives of Insect Biochemistry and Physiology 80:77–91. - PubMed
-
- Qin G, Jia M, Liu T, Xuan T, Zhu K, et al. (2011) Identification and characterisation of ten glutathione S-transferase genes from oriental migratory locust, Locusta migratoria manilensis (Meyen). Pest Management Science 67:697–704. - PubMed
-
- Li X, Zhang X, Zhang J, Zhang X, Starkey SR, et al. (2009) Identification and characterization of eleven glutathione S-transferase genes from the aquatic midge Chironomus tentans (Diptera: Chironomidae). Insect Biochemistry and Molecular Biology 39:745–754. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

