A mutation in Na(+)-NQR uncouples electron flow from Na(+) translocation in the presence of K(+)
- PMID: 25486106
- PMCID: PMC5019955
- DOI: 10.1021/bi501266e
A mutation in Na(+)-NQR uncouples electron flow from Na(+) translocation in the presence of K(+)
Abstract
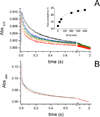
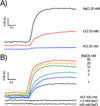
The sodium-pumping NADH:ubiquinone oxidoreductase (Na(+)-NQR) is a bacterial respiratory enzyme that obtains energy from the redox reaction between NADH and ubiquinone and uses this energy to create an electrochemical Na(+) gradient across the cell membrane. A number of acidic residues in transmembrane helices have been shown to be important for Na(+) translocation. One of these, Asp-397 in the NqrB subunit, is a key residue for Na(+) uptake and binding. In this study, we show that when this residue is replaced with asparagine, the enzyme acquires a new sensitivity to K(+); in the mutant, K(+) both activates the redox reaction and uncouples it from the ion translocation reaction. In the wild-type enzyme, Na(+) (or Li(+)) accelerates turnover while K(+) alone does not activate. In the NqrB-D397N mutant, K(+) accelerates the same internal electron transfer step (2Fe-2S → FMNC) that is accelerated by Na(+). This is the same step that is inhibited in mutants in which Na(+) uptake is blocked. NqrB-D397N is able to translocate Na(+) and Li(+), but when K(+) is introduced, no ion translocation is observed, regardless of whether Na(+) or Li(+) is present. Thus, this mutant, when it turns over in the presence of K(+), is the first, and currently the only, example of an uncoupled Na(+)-NQR. The fact the redox reaction and ion pumping become decoupled from each other only in the presence of K(+) provides a switch that promises to be a useful experimental tool.
Figures


Similar articles
-
Glutamate 95 in NqrE Is an Essential Residue for the Translocation of Cations in Na+-NQR.Biochemistry. 2019 Apr 23;58(16):2167-2175. doi: 10.1021/acs.biochem.8b01294. Epub 2019 Apr 8. Biochemistry. 2019. PMID: 30907577
-
The role of glycine residues 140 and 141 of subunit B in the functional ubiquinone binding site of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae.J Biol Chem. 2012 Jul 20;287(30):25678-85. doi: 10.1074/jbc.M112.366088. Epub 2012 May 29. J Biol Chem. 2012. PMID: 22645140 Free PMC article.
-
Purification and characterization of the recombinant Na(+)-translocating NADH:quinone oxidoreductase from Vibrio cholerae.Biochemistry. 2002 Mar 19;41(11):3781-9. doi: 10.1021/bi011873o. Biochemistry. 2002. PMID: 11888296
-
The Na+-translocating NADH:quinone oxidoreductase (Na+-NQR): Physiological role, structure and function of a redox-driven, molecular machine.Biochim Biophys Acta Bioenerg. 2024 Nov 1;1865(4):149485. doi: 10.1016/j.bbabio.2024.149485. Epub 2024 Jun 30. Biochim Biophys Acta Bioenerg. 2024. PMID: 38955304 Review.
-
The structure of Na⁺-translocating of NADH:ubiquinone oxidoreductase of Vibrio cholerae: implications on coupling between electron transfer and Na⁺ transport.Biol Chem. 2015 Sep;396(9-10):1015-30. doi: 10.1515/hsz-2015-0128. Biol Chem. 2015. PMID: 26146127 Review.
Cited by
-
Identification of the Catalytic Ubiquinone-binding Site of Vibrio cholerae Sodium-dependent NADH Dehydrogenase: A NOVEL UBIQUINONE-BINDING MOTIF.J Biol Chem. 2017 Feb 17;292(7):3039-3048. doi: 10.1074/jbc.M116.770982. Epub 2017 Jan 4. J Biol Chem. 2017. PMID: 28053088 Free PMC article.
-
The Kinetic Reaction Mechanism of the Vibrio cholerae Sodium-dependent NADH Dehydrogenase.J Biol Chem. 2015 Aug 14;290(33):20009-21. doi: 10.1074/jbc.M115.658773. Epub 2015 May 23. J Biol Chem. 2015. PMID: 26004776 Free PMC article.
-
Characterization of the Pseudomonas aeruginosa NQR complex, a bacterial proton pump with roles in autopoisoning resistance.J Biol Chem. 2018 Oct 5;293(40):15664-15677. doi: 10.1074/jbc.RA118.003194. Epub 2018 Aug 22. J Biol Chem. 2018. PMID: 30135204 Free PMC article.
-
Metabolic mechanism of ceftazidime resistance in Vibrio alginolyticus.Infect Drug Resist. 2019 Feb 13;12:417-429. doi: 10.2147/IDR.S179639. eCollection 2019. Infect Drug Resist. 2019. PMID: 30863124 Free PMC article.
References
-
- Dibrov PA, Kostryko VA, Lazarova RL, Skulachev VP, Smirnova IA. The sodium cycle. I. Na+-dependent motility and modes of membrane energization in the marine alkalotolerant Vibrio alginolyticus. Biochim. Biophys. Acta. 1986;850:449–457. - PubMed
-
- Dibrov PA, Lazarova RL, Skulachev VP, Verkhovskaya ML. The sodium cycle. II. Na+-coupled oxidative phosphorylation in Vibrio alginolyticus cells. Biochim. Biophys. Acta. 1986;850:458–465. - PubMed
-
- Terashima H, Kojima S, Homma M. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 2008;270:39–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

