An entity evolving into a community: defining the common ancestor and evolutionary trajectory of chronic lymphocytic leukemia stereotyped subset #4
- PMID: 25486132
- PMCID: PMC4398667
- DOI: 10.2119/molmed.2014.00140
An entity evolving into a community: defining the common ancestor and evolutionary trajectory of chronic lymphocytic leukemia stereotyped subset #4
Abstract
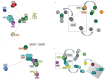
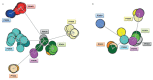
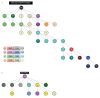
Patients with chronic lymphocytic leukemia (CLL) assigned to stereotyped subset #4 express highly homologous B-cell receptor immunoglobulin (BcR IG) sequences with intense intraclonal diversification (ID) in the context of ongoing somatic hypermutation (SHM). Their remarkable biological and clinical similarities strongly support derivation from a common ancestor. We here revisited ID in subset #4 CLL to reconstruct their evolutionary history as a community of related clones. To this end, using specialized bioinformatics tools we assessed both IGHV-IGHD-IGHJ rearrangements (n = 511) and IGKV-IGKJ rearrangements (n = 397) derived from eight subset #4 cases. Due to high sequence relatedness, a number of subclonal clusters from different cases lay very close to one another, forming a core from which clusters exhibiting greater variation stemmed. Minor subclones from individual cases were mutated to such an extent that they now resembled the sequences of another patient. Viewing the entire subset #4 data set as a single entity branching through diversification enabled inference of a common sequence representing the putative ancestral BcR IG expressed by their still elusive common progenitor. These results have implications for improved understanding of the ontogeny of CLL subset #4, as well as the design of studies concerning the antigenic specificity of the clonotypic BcR IGs.
Figures

Similar articles
-
Mutational status and gene repertoire of IGHV-IGHD-IGHJ rearrangements in Serbian patients with chronic lymphocytic leukemia.Clin Lymphoma Myeloma Leuk. 2012 Aug;12(4):252-60. doi: 10.1016/j.clml.2012.03.005. Epub 2012 May 4. Clin Lymphoma Myeloma Leuk. 2012. PMID: 22560084
-
Extensive intraclonal diversification in a subgroup of chronic lymphocytic leukemia patients with stereotyped IGHV4-34 receptors: implications for ongoing interactions with antigen.Blood. 2009 Nov 12;114(20):4460-8. doi: 10.1182/blood-2009-05-221309. Epub 2009 Aug 27. Blood. 2009. PMID: 19713457
-
Molecular and clinical features of chronic lymphocytic leukaemia with stereotyped B cell receptors: results from an Italian multicentre study.Br J Haematol. 2009 Feb;144(4):492-506. doi: 10.1111/j.1365-2141.2008.07469.x. Epub 2008 Nov 19. Br J Haematol. 2009. PMID: 19036101
-
Chronic lymphocytic leukaemia: an immunobiology approach.Srp Arh Celok Lek. 2008 May-Jun;136(5-6):319-23. doi: 10.2298/sarh0806319k. Srp Arh Celok Lek. 2008. PMID: 18792635 Review.
-
Immunogenetic studies of chronic lymphocytic leukemia: revelations and speculations about ontogeny and clinical evolution.Cancer Res. 2014 Aug 15;74(16):4211-6. doi: 10.1158/0008-5472.CAN-14-0630. Epub 2014 Jul 29. Cancer Res. 2014. PMID: 25074616 Review.
Cited by
-
Somatic hypermutation profiles in stereotyped IGHV4-34 receptors from South American chronic lymphocytic leukemia patients.Ann Hematol. 2022 Feb;101(2):341-348. doi: 10.1007/s00277-021-04703-9. Epub 2021 Oct 28. Ann Hematol. 2022. PMID: 34713310
-
Next-generation sequencing in chronic lymphocytic leukemia: recent findings and new horizons.Oncotarget. 2017 Jul 24;8(41):71234-71248. doi: 10.18632/oncotarget.19525. eCollection 2017 Sep 19. Oncotarget. 2017. PMID: 29050359 Free PMC article. Review.
-
Antigen receptor stereotypy in chronic lymphocytic leukemia.Leukemia. 2017 Feb;31(2):282-291. doi: 10.1038/leu.2016.322. Epub 2016 Nov 4. Leukemia. 2017. PMID: 27811850 Review.
References
-
- Stamatopoulos K, et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood. 2007;109:259–70. - PubMed
-
- Baliakas P, et al. Clinical effect of stereotyped B-cell receptor immunoglobulins in chronic lymphocytic leukaemia: a retrospective multicentre study. Lancet Haematol. 2014;1:e74–84. - PubMed
-
- Murray F, et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood. 2008;111:1524–33. - PubMed
-
- Vardi A, et al. IgG-switched CLL has a distinct immunogenetic signature from the common MD variant: ontogenetic implications. Clin Cancer Res. 2014;20:323–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

