Construction of synthetic nucleoli and what it tells us about propagation of sub-nuclear domains through cell division
- PMID: 25486191
- PMCID: PMC4614152
- DOI: 10.4161/15384101.2014.949124
Construction of synthetic nucleoli and what it tells us about propagation of sub-nuclear domains through cell division
Abstract
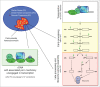
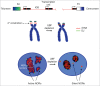
The cell nucleus is functionally compartmentalized into numerous membraneless and dynamic, yet defined, bodies. The cell cycle inheritance of these nuclear bodies (NBs) is poorly understood at the molecular level. In higher eukaryotes, their propagation is challenged by cell division through an "open" mitosis, where the nuclear envelope disassembles along with most NBs. A deeper understanding of the mechanisms involved can be achieved using the engineering principles of synthetic biology to construct artificial NBs. Successful biogenesis of such synthetic NBs demonstrates knowledge of the basic mechanisms involved. Application of this approach to the nucleolus, a paradigm of nuclear organization, has highlighted a key role for mitotic bookmarking in the cell cycle propagation of NBs.
Keywords: 1°, primary; 2°, secondary; CBs, Cajal bodies; CDK, cyclin-dependent kinase; DFC, dense fibrillar component; DJ, distal junction; FCs, fibrillar centers; GC, granular component; HLBs, histone locus bodies; HMG, high mobility group; IGS, intergenic spacers; NBs, nuclear bodies; NORs, nucleolar organizer regions; Nucleolar Organizer Region (NOR); PJ, proximal junction; PML, promyelocytic leukemia; PNBs, pre-nucleolar bodies; TFs, transcription factors; UBF; UBF, Upstream binding factor; XEn, Xenopus enhancer; cell cycle; mitotic bookmarking; neo-NOR; neonucleoli; nuclear bodies; nucleolus; pol, RNA polymerase; pre-rRNA, precursor rRNA; pseudo-NOR; rDNA, ribosomal genes; rRNA, ribosomal RNA; RNP, ribonucleoprotein; synthetic biology; t-UTPs, transcription U 3 proteins.
Figures

Similar articles
-
Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division.Genes Dev. 2014 Feb 1;28(3):220-30. doi: 10.1101/gad.234591.113. Epub 2014 Jan 21. Genes Dev. 2014. PMID: 24449107 Free PMC article.
-
The functional organization of the nucleolus in proliferating plant cells.Eur J Histochem. 2000;44(2):117-31. Eur J Histochem. 2000. PMID: 10968360 Review.
-
The nucleolus.Anat Embryol (Berl). 1993 Dec;188(6):515-36. doi: 10.1007/BF00187008. Anat Embryol (Berl). 1993. PMID: 8129175 Review.
-
The traffic of proteins between nucleolar organizer regions and prenucleolar bodies governs the assembly of the nucleolus at exit of mitosis.Nucleus. 2010 Mar-Apr;1(2):202-11. doi: 10.4161/nucl.1.2.11334. Epub 2010 Jan 28. Nucleus. 2010. PMID: 21326952 Free PMC article.
-
Driving nucleolar assembly.Genes Dev. 2014 Feb 1;28(3):211-3. doi: 10.1101/gad.237610.114. Genes Dev. 2014. PMID: 24493643 Free PMC article.
Cited by
-
Che-1/AATF binds to RNA polymerase I machinery and sustains ribosomal RNA gene transcription.Nucleic Acids Res. 2020 Jun 19;48(11):5891-5906. doi: 10.1093/nar/gkaa344. Nucleic Acids Res. 2020. PMID: 32421830 Free PMC article.
-
Trnp1 organizes diverse nuclear membrane-less compartments in neural stem cells.EMBO J. 2020 Aug 17;39(16):e103373. doi: 10.15252/embj.2019103373. Epub 2020 Jul 6. EMBO J. 2020. PMID: 32627867 Free PMC article.
-
Determinants of mammalian nucleolar architecture.Chromosoma. 2015 Sep;124(3):323-31. doi: 10.1007/s00412-015-0507-z. Epub 2015 Feb 12. Chromosoma. 2015. PMID: 25670395 Free PMC article. Review.
-
Super-resolution in situ analysis of active ribosomal DNA chromatin organization in the nucleolus.Sci Rep. 2020 May 4;10(1):7462. doi: 10.1038/s41598-020-64589-x. Sci Rep. 2020. PMID: 32366902 Free PMC article.
-
Close to the edge: Heterochromatin at the nucleolar and nuclear peripheries.Biochim Biophys Acta Gene Regul Mech. 2021 Jan;1864(1):194666. doi: 10.1016/j.bbagrm.2020.194666. Epub 2020 Dec 8. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 33307247 Free PMC article. Review.
References
-
- Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspec Biol 2010; 2:1-5; http://dx.doi.org/10.1101/cshperspect.a000711 - DOI - PMC - PubMed
-
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochemcell Biol 2008; 129:13-31; PMID:18046571; http://dx.doi.org/10.1007/s00418-007-0359-6 - DOI - PMC - PubMed
-
- Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, Melino G, De Laurenzi V. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle 2008; 7:2357-67; PMID:18677100; http://dx.doi.org/10.4161/cc.6344 - DOI - PubMed
-
- Platani M, Goldberg I, Lamond AI, Swedlow JR. Cajal body dynamics and association with chromatin are ATP-dependent. Nat Cell Ciol 2002; 4:502-8; PMID:12068306; http://dx.doi.org/10.1038/ncb809 - DOI - PubMed
-
- Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene 2002; 21:598-610; PMID:11850785; http://dx.doi.org/10.1038/sj.onc.1205058 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
