CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells
- PMID: 25486255
- PMCID: PMC4259346
- DOI: 10.1371/journal.pone.0114485
CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells
Abstract
The pluripotency of embryonic stem cells (ESCs) is maintained by a small group of master transcription factors including Oct4, Sox2 and Nanog. These core factors form a regulatory circuit controlling the transcription of a number of pluripotency factors including themselves. Although previous studies have identified transcriptional regulators of this core network, the cis-regulatory DNA sequences required for the transcription of these key pluripotency factors remain to be defined. We analyzed epigenomic data within the 1.5 Mb gene-desert regions around the Sox2 gene and identified a 13kb-long super-enhancer (SE) located 100kb downstream of Sox2 in mouse ESCs. This SE is occupied by Oct4, Sox2, Nanog, and the mediator complex, and physically interacts with the Sox2 locus via DNA looping. Using a simple and highly efficient double-CRISPR genome editing strategy we deleted the entire 13-kb SE and characterized transcriptional defects in the resulting monoallelic and biallelic deletion clones with RNA-seq. We showed that the SE is responsible for over 90% of Sox2 expression, and Sox2 is the only target gene along the chromosome. Our results support the functional significance of a SE in maintaining the pluripotency transcription program in mouse ESCs.
Conflict of interest statement
Figures
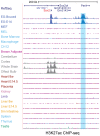
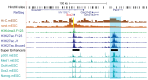

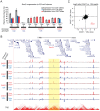
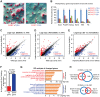
References
-
- Chen X, Xu H, Yuan P, Fang F, Huss M, et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133:1106–1117. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

