The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release
- PMID: 25486306
- PMCID: PMC4286497
- DOI: 10.1038/nsmb.2930
The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release
Abstract
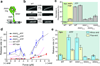
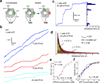
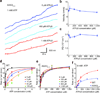
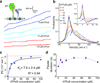
Cytoplasmic dynein is an AAA+ motor responsible for intracellular cargo transport and force generation along microtubules (MTs). Unlike kinesin and myosin, dynein contains multiple ATPase subunits, with AAA1 serving as the primary catalytic site. ATPase activity at AAA3 is also essential for robust motility, but its role in dynein's mechanochemical cycle remains unclear. Here, we introduced transient pauses in Saccharomyces cerevisiae dynein motility by using a slowly hydrolyzing ATP analog. Analysis of pausing behavior revealed that AAA3 hydrolyzes nucleotide an order of magnitude more slowly than AAA1, and the two sites do not coordinate. ATPase mutations to AAA3 abolish the ability of dynein to modulate MT release. Nucleotide hydrolysis at AAA3 lifts this 'MT gate' to allow fast motility. These results suggest that AAA3 acts as a switch that repurposes cytoplasmic dynein for fast cargo transport and MT-anchoring tasks in cells.
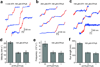
Figures


References
-
- Stokin GB, Goldstein LSB. Axonal transport and Alzheimer’s disease. Annu. Rev. Biochem. 2006;75:607–627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

