Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond
- PMID: 25486472
- PMCID: PMC4614920
- DOI: 10.4161/15384101.2014.949201
Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond
Abstract
Many biologically significant processes, such as cell differentiation and cell cycle progression, gene transcription and DNA replication, chromosome stability and epigenetic silencing etc. depend on the crucial interactions between cellular proteins and DNA. Chromatin immunoprecipitation (ChIP) is an important experimental technique for studying interactions between specific proteins and DNA in the cell and determining their localization on a specific genomic locus. In recent years, the combination of ChIP with second generation DNA-sequencing technology (ChIP-seq) allows precise genomic functional assay. This review addresses the important applications of ChIP-seq with an emphasis on its role in genome-wide mapping of transcription factor binding sites, the revelation of underlying molecular mechanisms of differential gene regulation that are governed by specific transcription factors, and the identification of epigenetic marks. Furthermore, we also describe the ChIP-seq data analysis workflow and a perspective for the exciting potential advancement of ChIP-seq technology in the future.
Keywords: AR; Burrows-Wheeler aligner; C. elegans; Caenorhabditis elegans; ChIP; ChIP sequencing; DNA; ChIP-seq processing pipeline; STAT; Model Organism ENCyclopedia Of DNA Elements; NF-κB; University of California Santa Cruz; UV; androgen receptor; BWA; c-Jun NH2-terminal kinase; K; chromatin; chromatin immunoprecipitation; CRs; chromatin regulators; ChIP-seq; deoxyribonuclease; EMSA; deoxyribonucleic acid; DNase; electrophoresis mobility shift assay; ENCODE; encyclopedia of DNA elements; FDR; false discovery rate; GR; glucocorticoid receptor; HDAC; haematopoietic stem progenitor cells; HM; high throughput; high throughput ChIP; IL-β; histone deacetylase; HEK; histone modification; HTChIP; human embryonic kidney; HSPCs; immunoprecipitation; interferon; JNK; interleukin β; IFN; lysine; MACS; model-based analysis of ChIP-seq; MEME; multiple Em for motif elicitation; modENCODE; nuclear factor κB; PCR; polymerase chain reaction; RNA; ribonucleic acid; R-ChIP; robotic ChIP; SNP; sequencing; short oligonucleotide alignment program; SPP; signal transducers and activators of transcription; SUMO; single nucleotide polymorphism; SOAP; small ubiquitin-like modifier; TFs; transcription factor binding sites; TSS; transcription factors; TFBS; transcription start site; UCSC; ultraviolet..
Figures
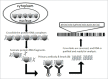
References
-
- Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-κB gene network by chromatin immunoprecipitation. Biotechniques 2005; 39:715-25; PMID:16315372; http://dx.doi.org/10.1007/978-1-61779-376-9_7 - DOI - PubMed
-
- Weinmann SA, Bartley MS, Zhang T, Zhang QM, Farnham JP. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell Biol 2001; 21:6820-32; PMID:11564866; http://dx.doi.org/10.1128/MCB.21.20.6820-6832.2001 - DOI - PMC - PubMed
-
- Weinmann SA, Farnham JP. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 2002; 26:37-47; PMID:12054903; http://dx.doi.org/10.1016/S1046-2023(02)00006-3 - DOI - PubMed
-
- Weinmann SA, Yan SP, Oberley JM, Huang HT, Farnham JP. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev 2002; 16:235-44; PMID:11799066; http://dx.doi.org/10.1101/gad.943102 - DOI - PMC - PubMed
-
- Spencer AV, Sun MJ, Li L, Davie RJ. Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods 2003; 31:67-5; PMID:12893175; http://dx.doi.org/10.1016/S1046-2023(03)00089-6 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
