ISWI chromatin remodeling complexes in the DNA damage response
- PMID: 25486562
- PMCID: PMC4615051
- DOI: 10.4161/15384101.2014.956551
ISWI chromatin remodeling complexes in the DNA damage response
Abstract
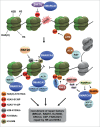
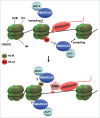
Regulation of chromatin structure is an essential component of the DNA damage response (DDR), which effectively preserves the integrity of DNA by a network of multiple DNA repair and associated signaling pathways. Within the DDR, chromatin is modified and remodeled to facilitate efficient DNA access, to control the activity of repair proteins and to mediate signaling. The mammalian ISWI family has recently emerged as one of the major ATP-dependent chromatin remodeling complex families that function in the DDR, as it is implicated in at least 3 major DNA repair pathways: homologous recombination, non-homologous end-joining and nucleotide excision repair. In this review, we discuss the various manners through which different ISWI complexes regulate DNA repair and how they are targeted to chromatin containing damaged DNA.
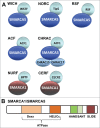
Keywords: ACF1; ACF1, ATP-utilizing Chromatin assembly and remodeling Factor 1; ATP-dependent chromatin remodeling; BER, Base Excision Repair; DDR, DNA Damage Response; DNA damage response; DSB, Double Strand Break; GG-NER, Global Genome Nucleotide Excision Repair; HR, Homologous Recombination; Homologous Recombination; ISWI; ISWI, Imitation SWItch; MRN, MRE11/Rad50/NBS1; NER, Nucleotide Excision Repair; NHEJ, Non-Homologous End Joining; Non-Homologous End-Joining; Nucleotide Excision Repair; PAR, Poly(ADP-Ribose); RNApolII, RNA Polymerase II; RSF1, Remodeling and Spacing Factor 1; SMARCA, SWI-SNF-related Matrix-associated Actin-dependent Regulator of Chromatin A; SMARCA5/SNF2H; TC-NER, Transcription-Coupled Nucleotide Excision Repair; WSTF; WSTF, Williams Syndrome Transcription Factor.
Figures

References
-
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med 2009; 361:1475-85; PMID:19812404; http://dx.doi.org/10.1056/NEJMra0804615 - DOI - PubMed
-
- Giglia-Mari G, Zotter A, Vermeulen W. DNA damage response. Cold Spring Harb Perspect Biol 2011; 3:a000745; PMID:20980439; http://dx.doi.org/10.1101/cshperspect.a000745 - DOI - PMC - PubMed
-
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013; 153:17-37; PMID:23540688; http://dx.doi.org/10.1016/j.cell.2013.03.002 - DOI - PubMed
-
- O’Sullivan RJ, Karlseder J. The great unravelling: chromatin as a modulator of the aging process. Trends Biochem Sci 2012; 37:466-76; http://dx.doi.org/10.1016/j.tibs.2012.08.001 - DOI - PMC - PubMed
-
- Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol 2012; 13:436-47; PMID:22722606; http://dx.doi.org/10.1038/nrm3382 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
