PML represses lung cancer metastasis by suppressing the nuclear EGFR-mediated transcriptional activation of MMP2
- PMID: 25486572
- PMCID: PMC4614437
- DOI: 10.4161/15384101.2014.949212
PML represses lung cancer metastasis by suppressing the nuclear EGFR-mediated transcriptional activation of MMP2
Abstract
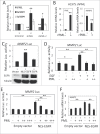
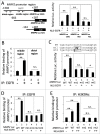
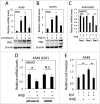
Promyelocytic leukemia protein (PML) is emerging as an important tumor suppressor. Its expression is lost during the progression of several types of cancer, including lung cancer. The EGF receptor (EGFR), a membrane-bound receptor tyrosine kinase, transduces intracellular signals responsible for cell proliferation, differentiation and migration. EGFR activity is frequently abnormally upregulated in lung adenocarcinoma (LAC) and thus is considered to be a driving oncogene for LAC. EGFR translocates into the nucleus and transcriptionally activates genes, such as CCND1, that promote cell growth. Recently, we demonstrated that PML interacted with nuclear EGFR (nEGFR) and suppressed the nEGFR-mediated transcriptional activation of CCND1 in lung cancer cells, thereby restraining cell growth. When we further investigated the interplay between PML and EGFR in lung cancer metastasis, we found that the matrix metalloprotease-2 gene (MMP2) was a novel nEGFR target gene and was repressed by PML. We provide evidence that nEGFR bound to the AT-rich sequence (ATRS) in the MMP2 promoter and enhanced its transcriptional activity. In addition, we demonstrated that PML repressed nEGFR-induced MMP2 transcription and reduced cell invasion. PML was recruited by nEGFR to the MMP2 promoter where it reduced histone acetylation, leading to the transcriptional repression of MMP2. Finally, we demonstrated that PML upregulation by interferon-β (IFNβ) in lung cancer cells decreased MMP2 expression and cell invasion. Together, our results suggested that IFNβ induced PML to inhibit lung cancer metastasis by repressing the nEGFR-mediated transcriptional activation of MMP2.
Keywords: ATRS; ATRS, AT-rich sequence; EGFR, EGF receptor; IFNβ, interferon-β; INM, inner nuclear membrane; Interferon-β; LAC, lung adenocarcinoma; MMP2; MMP2, matrix metalloprotease-2; PML; PML, promyelocytic leukemia protein; STAT3; lung adenocarcinoma; nEGFR, nuclear EGFR; nuclear EGFR.
Figures



Similar articles
-
The PML isoform IV is a negative regulator of nuclear EGFR's transcriptional activity in lung cancer.Carcinogenesis. 2013 Aug;34(8):1708-16. doi: 10.1093/carcin/bgt109. Epub 2013 Apr 4. Carcinogenesis. 2013. PMID: 23563092
-
PGE2/EP3/SRC signaling induces EGFR nuclear translocation and growth through EGFR ligands release in lung adenocarcinoma cells.Oncotarget. 2017 May 9;8(19):31270-31287. doi: 10.18632/oncotarget.16116. Oncotarget. 2017. PMID: 28415726 Free PMC article.
-
Direct transcriptional activation of promyelocytic leukemia protein by IFN regulatory factor 3 induces the p53-dependent growth inhibition of cancer cells.Cancer Res. 2007 Dec 1;67(23):11133-40. doi: 10.1158/0008-5472.CAN-07-1342. Cancer Res. 2007. PMID: 18056437
-
[Promyelocytic leukaemia protein and defect in transforming growth factor-beta signal pathway in acute promyelocytic leukaemia].Cas Lek Cesk. 2005;144(2):90-4. Cas Lek Cesk. 2005. PMID: 15807293 Review. Czech.
-
Novel treatment of acute promyelocytic leukemia: As₂O₃, retinoic acid and retinoid pharmacology.Curr Pharm Biotechnol. 2013;14(9):849-58. doi: 10.2174/1389201015666140113095812. Curr Pharm Biotechnol. 2013. PMID: 24433507 Review.
Cited by
-
Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases.FEBS J. 2015 Oct;282(19):3693-721. doi: 10.1111/febs.13342. Epub 2015 Jul 4. FEBS J. 2015. PMID: 26096795 Free PMC article. Review.
-
High expression of RUNX1 is associated with poorer outcomes in cytogenetically normal acute myeloid leukemia.Oncotarget. 2016 Mar 29;7(13):15828-39. doi: 10.18632/oncotarget.7489. Oncotarget. 2016. PMID: 26910834 Free PMC article.
-
mRNA Expression Analysis of E-Cadherin, VEGF, and MMPs in Gastric Cancer: a Pilot Study.Indian J Surg Oncol. 2021 Apr;12(Suppl 1):85-92. doi: 10.1007/s13193-020-01096-5. Epub 2020 May 22. Indian J Surg Oncol. 2021. PMID: 33994733 Free PMC article.
-
Epigenetic silencing of tumor suppressor candidate 3 confers adverse prognosis in early colorectal cancer.Oncotarget. 2017 Sep 15;8(49):84714-84728. doi: 10.18632/oncotarget.20950. eCollection 2017 Oct 17. Oncotarget. 2017. PMID: 29156678 Free PMC article.
-
Advances in the role of membrane-bound transcription factors in carcinogenesis and therapy.Discov Oncol. 2024 Oct 15;15(1):559. doi: 10.1007/s12672-024-01414-1. Discov Oncol. 2024. PMID: 39404930 Free PMC article. Review.
References
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69-90; PMID:21296855; http://dx.doi.org/10.3322/caac.20107 - DOI - PubMed
-
- Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 2006; 12:895-904; PMID: 16892035; http://dx.doi.org/10.1038/nm1469 - DOI - PubMed
-
- Guo CB, Wang S, Deng C, Zhang DL, Wang FL, Jin XQ. Relationship between matrix metalloproteinase 2 and lung cancer progression. Mol Diagn Ther 2007; 11:183-92; PMID:17570740; http://dx.doi.org/10.1007/BF03256240 - DOI - PubMed
-
- Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev 2006; 25:9-34; PMID:16680569; http://dx.doi.org/10.1007/s10555-006-7886-9 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
