Fine structure of the vaccinia virion determined by controlled degradation and immunolocalization
- PMID: 25486587
- PMCID: PMC4280304
- DOI: 10.1016/j.virol.2014.11.020
Fine structure of the vaccinia virion determined by controlled degradation and immunolocalization
Abstract
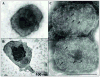
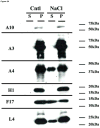
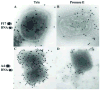
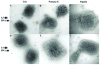
The vaccinia virion is a membraned, slightly flattened, barrel-shaped particle, with a complex internal structure featuring a biconcave core flanked by lateral bodies. Although the architecture of the purified mature virion has been intensely characterized by electron microscopy, the distribution of the proteins within the virion has been examined primarily using biochemical procedures. Thus, it has been shown that non-ionic and ionic detergents combined or not with a sulfhydryl reagent can be used to disrupt virions and, to a limited degree, separate the constituent proteins in different fractions. Applying a controlled degradation technique to virions adsorbed on EM grids, we were able to immuno-localize viral proteins within the virion particle. Our results show after NP40 and DTT treatment, membrane proteins are removed from the virion surface revealing proteins that are associated with the lateral bodies and the outer layer of the core wall. Combined treatment using high salt and high DTT removed lateral body proteins and exposed proteins of the internal core wall. Cores treated with proteases could be disrupted and the internal components were exposed. Cts8, a mutant in the A3 protein, produces aberrant virus that, when treated with NP-40 and DTT, releases to the exterior the virus DNA associated with other internal core proteins. With these results, we are able to propose a model for the structure the vaccinia virion.
Keywords: Controlled degradation; Immunolocalization of proteins; Vaccinia virus; Virion structure.
Published by Elsevier Inc.
Figures









References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley & Sons; 1994.
-
- Carter GC, Law M, Hollinshead M, Smith GL. Entry of the vaccinia virus intracellular mature virion and its interactions with glycosaminoglycans. J Gen Virol. 2005;86:1279–1290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

