Next-generation sequencing of flow-sorted wheat chromosome 5D reveals lineage-specific translocations and widespread gene duplications
- PMID: 25487001
- PMCID: PMC4298962
- DOI: 10.1186/1471-2164-15-1080
Next-generation sequencing of flow-sorted wheat chromosome 5D reveals lineage-specific translocations and widespread gene duplications
Abstract
Background: The ~17 Gb hexaploid bread wheat genome is a high priority and a major technical challenge for genomic studies. In particular, the D sub-genome is relatively lacking in genetic diversity, making it both difficult to map genetically, and a target for introgression of agriculturally useful traits. Elucidating its sequence and structure will therefore facilitate wheat breeding and crop improvement.
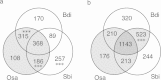
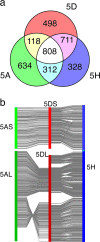
Results: We generated shotgun sequences from each arm of flow-sorted Triticum aestivum chromosome 5D using 454 FLX Titanium technology, giving 1.34× and 1.61× coverage of the short (5DS) and long (5DL) arms of the chromosome respectively. By a combination of sequence similarity and assembly-based methods, ~74% of the sequence reads were classified as repetitive elements, and coding sequence models of 1314 (5DS) and 2975 (5DL) genes were generated. The order of conserved genes in syntenic regions of previously sequenced grass genomes were integrated with physical and genetic map positions of 518 wheat markers to establish a virtual gene order for chromosome 5D.
Conclusions: The virtual gene order revealed a large-scale chromosomal rearrangement in the peri-centromeric region of 5DL, and a concentration of non-syntenic genes in the telomeric region of 5DS. Although our data support the large-scale conservation of Triticeae chromosome structure, they also suggest that some regions are evolving rapidly through frequent gene duplications and translocations.
Sequence accessions: EBI European Nucleotide Archive, Study no. ERP002330.
Figures





References
-
- Gill BS, Appels R, Botha-Oberholster A-M, Buell CR, Bennetzen JL, Chalhoub B, Chumley F, Dvorák J, Iwanaga M, Keller B, Li W, McCombie WR, Ogihara Y, Quetier F, Sasaki T. A workshop report on wheat genome sequencing: ınternational genome research on wheat consortium. Genetics. 2004;168:1087–1096. doi: 10.1534/genetics.104.034769. - DOI - PMC - PubMed
-
- Peng JH, Sun D, Nevo E. Domestication evolution, genetics and genomics in wheat. Mol Breed. 2011;28:281–301. doi: 10.1007/s11032-011-9608-4. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

