Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer
- PMID: 25487147
- PMCID: PMC4359913
- DOI: 10.1038/nature13996
Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer
Abstract
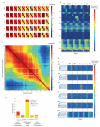
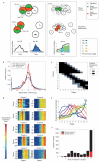
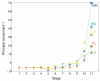
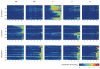
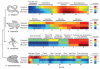
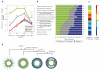
The concept of germ layers has been one of the foremost organizing principles in developmental biology, classification, systematics and evolution for 150 years (refs 1 - 3). Of the three germ layers, the mesoderm is found in bilaterian animals but is absent in species in the phyla Cnidaria and Ctenophora, which has been taken as evidence that the mesoderm was the final germ layer to evolve. The origin of the ectoderm and endoderm germ layers, however, remains unclear, with models supporting the antecedence of each as well as a simultaneous origin. Here we determine the temporal and spatial components of gene expression spanning embryonic development for all Caenorhabditis elegans genes and use it to determine the evolutionary ages of the germ layers. The gene expression program of the mesoderm is induced after those of the ectoderm and endoderm, thus making it the last germ layer both to evolve and to develop. Strikingly, the C. elegans endoderm and ectoderm expression programs do not co-induce; rather the endoderm activates earlier, and this is also observed in the expression of endoderm orthologues during the embryology of the frog Xenopus tropicalis, the sea anemone Nematostella vectensis and the sponge Amphimedon queenslandica. Querying the phylogenetic ages of specifically expressed genes reveals that the endoderm comprises older genes. Taken together, we propose that the endoderm program dates back to the origin of multicellularity, whereas the ectoderm originated as a secondary germ layer freed from ancestral feeding functions.
Figures





References
-
- Hall BK. Evolutionary developmental biology. 2nd ed. Chapman & Hall; 1998.
-
- Wolpert L. Principles of development. 4th ed. Oxford University Press; 2011.
-
- Technau U, Scholz CB. Origin and evolution of endoderm and mesoderm. Int J Dev Biol. 2003;47:531–539. - PubMed
-
- Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa) Development. 2004;131:2463–2474. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

