Novel dengue virus NS2B/NS3 protease inhibitors
- PMID: 25487800
- PMCID: PMC4335830
- DOI: 10.1128/AAC.03543-14
Novel dengue virus NS2B/NS3 protease inhibitors
Abstract
Dengue fever is a severe, widespread, and neglected disease with more than 2 million diagnosed infections per year. The dengue virus NS2B/NS3 protease (PR) represents a prime target for rational drug design. At the moment, there are no clinical PR inhibitors (PIs) available. We have identified diaryl (thio)ethers as candidates for a novel class of PIs. Here, we report the selective and noncompetitive inhibition of the serotype 2 and 3 dengue virus PR in vitro and in cells by benzothiazole derivatives exhibiting 50% inhibitory concentrations (IC50s) in the low-micromolar range. Inhibition of replication of DENV serotypes 1 to 3 was specific, since all substances influenced neither hepatitis C virus (HCV) nor HIV-1 replication. Molecular docking suggests binding at a specific allosteric binding site. In addition to the in vitro assays, a cell-based PR assay was developed to test these substances in a replication-independent way. The new compounds inhibited the DENV PR with IC50s in the low-micromolar or submicromolar range in cells. Furthermore, these novel PIs inhibit viral replication at submicromolar concentrations.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

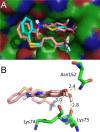


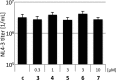

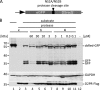

References
-
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi:10.1038/nature12060. - DOI - PMC - PubMed
-
- World Health Organization. November 2012, posting date Dengue and severe dengue. Fact sheet no. 117 World Health Organization, Geneva, Switzerland.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

