Role of the Streptococcus mutans CRISPR-Cas systems in immunity and cell physiology
- PMID: 25488301
- PMCID: PMC4334182
- DOI: 10.1128/JB.02333-14
Role of the Streptococcus mutans CRISPR-Cas systems in immunity and cell physiology
Abstract
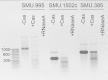
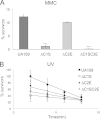
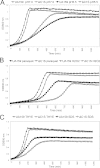
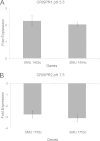
CRISPR-Cas systems provide adaptive microbial immunity against invading viruses and plasmids. The cariogenic bacterium Streptococcus mutans UA159 has two CRISPR-Cas systems: CRISPR1 (type II-A) and CRISPR2 (type I-C) with several spacers from both CRISPR cassettes matching sequences of phage M102 or genomic sequences of other S. mutans. The deletion of the cas genes of CRISPR1 (ΔC1S), CRISPR2 (ΔC2E), or both CRISPR1+2 (ΔC1SC2E) or the removal of spacers 2 and 3 (ΔCR1SP13E) in S. mutans UA159 did not affect phage sensitivity when challenged with virulent phage M102. Using plasmid transformation experiments, we demonstrated that the CRISPR1-Cas system inhibits transformation of S. mutans by the plasmids matching the spacers 2 and 3. Functional analysis of the cas deletion mutants revealed that in addition to a role in plasmid targeting, both CRISPR systems also contribute to the regulation of bacterial physiology in S. mutans. Compared to wild-type cells, the ΔC1S strain displayed diminished growth under cell membrane and oxidative stress, enhanced growth under low pH, and had reduced survival under heat shock and DNA-damaging conditions, whereas the ΔC2E strain exhibited increased sensitivity to heat shock. Transcriptional analysis revealed that the two-component signal transduction system VicR/K differentially modulates expression of cas genes within CRISPR-Cas systems, suggesting that VicR/K might coordinate the expression of two CRISPR-Cas systems. Collectively, we provide in vivo evidence that the type II-A CRISPR-Cas system of S. mutans may be targeted to manipulate its stress response and to influence the host to control the uptake and dissemination of antibiotic resistance genes.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

