A chromosomally encoded T7 RNA polymerase-dependent gene expression system for Corynebacterium glutamicum: construction and comparative evaluation at the single-cell level
- PMID: 25488698
- PMCID: PMC4353339
- DOI: 10.1111/1751-7915.12236
A chromosomally encoded T7 RNA polymerase-dependent gene expression system for Corynebacterium glutamicum: construction and comparative evaluation at the single-cell level
Abstract
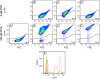
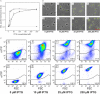
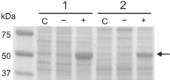
Corynebacterium glutamicum has become a favourite model organism in white biotechnology. Nevertheless, only few systems for the regulatable (over)expression of homologous and heterologous genes are currently available, all of which are based on the endogenous RNA polymerase. In this study, we developed an isopropyl-β-D-1-thiogalactopyranosid (IPTG)-inducible T7 expression system in the prophage-free strain C. glutamicum MB001. For this purpose, part of the DE3 region of Escherichia coli BL21(DE3) including the T7 RNA polymerase gene 1 under control of the lacUV5 promoter was integrated into the chromosome, resulting in strain MB001(DE3). Furthermore, the expression vector pMKEx2 was constructed allowing cloning of target genes under the control of the T7lac promoter. The properties of the system were evaluated using eyfp as heterologous target gene. Without induction, the system was tightly repressed, resulting in a very low specific eYFP fluorescence (= fluorescence per cell density). After maximal induction with IPTG, the specific fluorescence increased 450-fold compared with the uninduced state and was about 3.5 times higher than in control strains expressing eyfp under control of the IPTG-induced tac promoter with the endogenous RNA polymerase. Flow cytometry revealed that T7-based eyfp expression resulted in a highly uniform population, with 99% of all cells showing high fluorescence. Besides eyfp, the functionality of the corynebacterial T7 expression system was also successfully demonstrated by overexpression of the C. glutamicum pyk gene for pyruvate kinase, which led to an increase of the specific activity from 2.6 to 135 U mg(-1). It thus presents an efficient new tool for protein overproduction, metabolic engineering and synthetic biology approaches with C. glutamicum.
© 2014 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures


References
-
- Arvani S, Markert A, Loeschcke A, Jaeger KE. Drepper T. A T7 RNA polymerase-based toolkit for the concerted expression of clustered genes. J Biotechnol. 2012;159:162–171. - PubMed
-
- Baneyx F. Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol. 1999;10:411–421. - PubMed
-
- Barnard GC, Henderson GE, Srinivasan S. Gerngross TU. High level recombinant protein expression in Ralstonia eutropha using T7 RNA polymerase based amplification. Protein Expr Purif. 2004;38:264–271. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

