Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module
- PMID: 25488812
- PMCID: PMC4288179
- DOI: 10.1093/nar/gku1287
Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module
Abstract
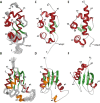
The La-related proteins (LARPs) form a diverse group of RNA-binding proteins characterized by the possession of a composite RNA binding unit, the La module. The La module comprises two domains, the La motif (LaM) and the RRM1, which together recognize and bind to a wide array of RNA substrates. Structural information regarding the La module is at present restricted to the prototypic La protein, which acts as an RNA chaperone binding to 3' UUUOH sequences of nascent RNA polymerase III transcripts. In contrast, LARP6 is implicated in the regulation of collagen synthesis and interacts with a specific stem-loop within the 5' UTR of the collagen mRNA. Here, we present the structure of the LaM and RRM1 of human LARP6 uncovering in both cases considerable structural variation in comparison to the equivalent domains in La and revealing an unprecedented fold for the RRM1. A mutagenic study guided by the structures revealed that RNA recognition requires synergy between the LaM and RRM1 as well as the participation of the interdomain linker, probably in realizing tandem domain configurations and dynamics required for substrate selectivity. Our study highlights a considerable complexity and plasticity in the architecture of the La module within LARPs.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Characterization of Sequence-Specific Binding of LARP6 to the 5' Stem-Loop of Type I Collagen mRNAs and Implications for Rational Design of Antifibrotic Drugs.J Mol Biol. 2022 Jan 30;434(2):167394. doi: 10.1016/j.jmb.2021.167394. Epub 2021 Dec 8. J Mol Biol. 2022. PMID: 34896113 Free PMC article.
-
(1)H, (15)N and (13)C chemical shift assignments of the La motif and RRM1 from human LARP6.Biomol NMR Assign. 2015 Oct;9(2):337-40. doi: 10.1007/s12104-015-9605-3. Epub 2015 Apr 22. Biomol NMR Assign. 2015. PMID: 25896032 Free PMC article.
-
The LARP6 La module from Tetrabaena socialis reveals structural and functional differences from plant and animal LARP6 homologues.RNA Biol. 2025 Dec;22(1):1-9. doi: 10.1080/15476286.2025.2489303. Epub 2025 Apr 9. RNA Biol. 2025. PMID: 40181506 Free PMC article.
-
Conserved and divergent features of the structure and function of La and La-related proteins (LARPs).Biochim Biophys Acta. 2010 May-Jun;1799(5-6):365-78. doi: 10.1016/j.bbagrm.2010.01.011. Epub 2010 Feb 2. Biochim Biophys Acta. 2010. PMID: 20138158 Free PMC article. Review.
-
LARP1 and LARP4: up close with PABP for mRNA 3' poly(A) protection and stabilization.RNA Biol. 2021 Feb;18(2):259-274. doi: 10.1080/15476286.2020.1868753. Epub 2021 Jan 31. RNA Biol. 2021. PMID: 33522422 Free PMC article. Review.
Cited by
-
1H, 13C, and 15N resonance assignment of the 5'SL-bound La domain of the human La-related protein 6.Biomol NMR Assign. 2025 Jun;19(1):165-173. doi: 10.1007/s12104-025-10232-7. Epub 2025 Apr 30. Biomol NMR Assign. 2025. PMID: 40304844
-
Noncanonical RNA binding of human La-related protein 6.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf682. doi: 10.1093/nar/gkaf682. Nucleic Acids Res. 2025. PMID: 40705921 Free PMC article.
-
1H, 13C, and 15N resonance assignments of the La Motif of the human La-related protein 1.Biomol NMR Assign. 2024 Jun;18(1):111-118. doi: 10.1007/s12104-024-10176-4. Epub 2024 May 1. Biomol NMR Assign. 2024. PMID: 38691336 Free PMC article.
-
Maternal Larp6 controls oocyte development, chorion formation and elevation.Development. 2020 Feb 26;147(4):dev187385. doi: 10.1242/dev.187385. Development. 2020. PMID: 32054660 Free PMC article.
-
TOP mRNPs: Molecular Mechanisms and Principles of Regulation.Biomolecules. 2020 Jun 27;10(7):969. doi: 10.3390/biom10070969. Biomolecules. 2020. PMID: 32605040 Free PMC article. Review.
References
-
- Alfano C., Sanfelice D., Babon J., Kelly G., Jacks A., Curry S., Conte M.R. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004;11:323–329. - PubMed
-
- Wolin S.L., Cedervall T. The La protein. Annu. Rev. Biochem. 2002;71:375–403. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

