Genetic architecture of natural variation of telomere length in Arabidopsis thaliana
- PMID: 25488978
- PMCID: PMC4317667
- DOI: 10.1534/genetics.114.172163
Genetic architecture of natural variation of telomere length in Arabidopsis thaliana
Abstract
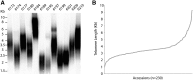
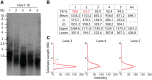
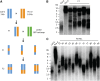
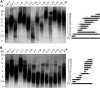
Telomeres represent the repetitive sequences that cap chromosome ends and are essential for their protection. Telomere length is known to be highly heritable and is derived from a homeostatic balance between telomeric lengthening and shortening activities. Specific loci that form the genetic framework underlying telomere length homeostasis, however, are not well understood. To investigate the extent of natural variation of telomere length in Arabidopsis thaliana, we examined 229 worldwide accessions by terminal restriction fragment analysis. The results showed a wide range of telomere lengths that are specific to individual accessions. To identify loci that are responsible for this variation, we adopted a quantitative trait loci (QTL) mapping approach with multiple recombinant inbred line (RIL) populations. A doubled haploid RIL population was first produced using centromere-mediated genome elimination between accessions with long (Pro-0) and intermediate (Col-0) telomere lengths. Composite interval mapping analysis of this population along with two established RIL populations (Ler-2/Cvi-0 and Est-1/Col-0) revealed a number of shared and unique QTL. QTL detected in the Ler-2/Cvi-0 population were examined using near isogenic lines that confirmed causative regions on chromosomes 1 and 2. In conclusion, this work describes the extent of natural variation of telomere length in A. thaliana, identifies a network of QTL that influence telomere length homeostasis, examines telomere length dynamics in plants with hybrid backgrounds, and shows the effects of two identified regions on telomere length regulation.
Keywords: Arabidopsis; QTL; centromere-mediated genome elimination; haploid; telomere.
Copyright © 2015 by the Genetics Society of America.
Figures



Similar articles
-
Components of the ribosome biogenesis pathway underlie establishment of telomere length set point in Arabidopsis.Nat Commun. 2019 Dec 2;10(1):5479. doi: 10.1038/s41467-019-13448-z. Nat Commun. 2019. PMID: 31792215 Free PMC article.
-
Rapid creation of Arabidopsis doubled haploid lines for quantitative trait locus mapping.Proc Natl Acad Sci U S A. 2012 Mar 13;109(11):4227-32. doi: 10.1073/pnas.1117277109. Epub 2012 Feb 27. Proc Natl Acad Sci U S A. 2012. PMID: 22371599 Free PMC article.
-
Identification of quantitative trait loci controlling symptom development during viral infection in Arabidopsis thaliana.Mol Plant Microbe Interact. 2008 Feb;21(2):198-207. doi: 10.1094/MPMI-21-2-0198. Mol Plant Microbe Interact. 2008. PMID: 18184064
-
Light-related loci controlling seed germination in Ler x Cvi and Bay-0 x Sha recombinant inbred-line populations of Arabidopsis thaliana.Ann Bot. 2008 Oct;102(4):631-42. doi: 10.1093/aob/mcn138. Epub 2008 Aug 6. Ann Bot. 2008. PMID: 18684732 Free PMC article.
-
Understanding genetic variation and function- the applications of next generation sequencing.Semin Cell Dev Biol. 2012 Apr;23(2):230-6. doi: 10.1016/j.semcdb.2012.01.006. Epub 2012 Jan 20. Semin Cell Dev Biol. 2012. PMID: 22285423 Review.
Cited by
-
Components of the ribosome biogenesis pathway underlie establishment of telomere length set point in Arabidopsis.Nat Commun. 2019 Dec 2;10(1):5479. doi: 10.1038/s41467-019-13448-z. Nat Commun. 2019. PMID: 31792215 Free PMC article.
-
Natural variation in plant telomere length is associated with flowering time.Plant Cell. 2021 May 31;33(4):1118-1134. doi: 10.1093/plcell/koab022. Plant Cell. 2021. PMID: 33580702 Free PMC article.
-
Fluorescent labelling of in situ hybridisation probes through the copper-catalysed azide-alkyne cycloaddition reaction.Chromosome Res. 2016 Sep;24(3):299-307. doi: 10.1007/s10577-016-9522-z. Epub 2016 Apr 19. Chromosome Res. 2016. PMID: 27095480
-
Arabidopsis telomerase takes off by uncoupling enzyme activity from telomere length maintenance in space.Nat Commun. 2023 Nov 29;14(1):7854. doi: 10.1038/s41467-023-41510-4. Nat Commun. 2023. PMID: 38030615 Free PMC article.
-
Telomeres are shorter in wild Saccharomyces cerevisiae isolates than in domesticated ones.Genetics. 2023 Mar 2;223(3):iyac186. doi: 10.1093/genetics/iyac186. Genetics. 2023. PMID: 36563016 Free PMC article.
References
-
- Alonso-Blanco C., Peeters A. J., Koornneef M., Lister C., Dean C., et al. , 1998b Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14: 259–271. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

