mtDNA Variation and Analysis Using Mitomap and Mitomaster
- PMID: 25489354
- PMCID: PMC4257604
- DOI: 10.1002/0471250953.bi0123s44
mtDNA Variation and Analysis Using Mitomap and Mitomaster
Abstract
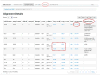
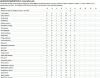
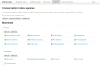
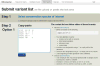
The Mitomap database of human mitochondrial DNA (mtDNA) information has been an important compilation of mtDNA variation for researchers, clinicians and genetic counselors for the past twenty-five years. The Mitomap protocol shows how users may look up human mitochondrial gene loci, search for public mitochondrial sequences, and browse or search for reported general population nucleotide variants as well as those reported in clinical disease. Within Mitomap is the powerful sequence analysis tool for human mitochondrial DNA, Mitomaster. The Mitomaster protocol gives step-by-step instructions showing how to submit sequences to identify nucleotide variants relative to the rCRS, to determine the haplogroup, and to view species conservation. User-supplied sequences, GenBank identifiers and single nucleotide variants may be analyzed.
Keywords: GenBank sequences; biological database; haplogroups; human mitochondrial DNA; information retrieval; single nucleotide variants; species conservation.
Figures









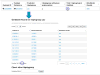
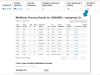

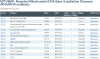
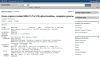



References
Literature Cited
-
- Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. - PubMed
-
- Kloss-Brandstatter A, Pacher D, Schonherr S, Weissensteiner H, Binna R, Specht G, Kronenberg F. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat. 2011;32:25–32. - PubMed
-
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat (Online) 2009;30:E386–E394. - PubMed
-
- Wallace DC, Lott MT, Brown MD, Huoponen K, Torroni A. Report of the committee on human mitochondrial DNA. In: Cuticchia AJ, editor. In Human Gene Mapping 1994, a Compendium. The Johns Hopkins University Press; Baltimore, MD: 1995. pp. 910–954.
Key References
-
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, Lehvaslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. Supporting publication for Bioperl. - PMC - PubMed
-
- Steinbiss S, Gremme G, Scharfer C, Mader M, Kurtz S. AnnotationSketch: a genome annotation drawing library. Bioinformatics. 2009;25:533–534. Supporting publication for AnnotationSketch. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

