Resolving cancer-stroma interfacial signalling and interventions with micropatterned tumour-stromal assays
- PMID: 25489927
- PMCID: PMC4261930
- DOI: 10.1038/ncomms6662
Resolving cancer-stroma interfacial signalling and interventions with micropatterned tumour-stromal assays
Abstract
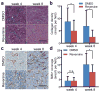
Tumour-stromal interactions are a determining factor in cancer progression. In vivo, the interaction interface is associated with spatially resolved distributions of cancer and stromal phenotypes. Here, we establish a micropatterned tumour-stromal assay (μTSA) with laser capture microdissection to control the location of co-cultured cells and analyse bulk and interfacial tumour-stromal signalling in driving cancer progression. μTSA reveals a spatial distribution of phenotypes in concordance with human oestrogen receptor-positive (ER+) breast cancer samples, and heterogeneous drug activity relative to the tumour-stroma interface. Specifically, an unknown mechanism of reversine is shown in targeting tumour-stromal interfacial interactions using ER+ MCF-7 breast cancer and bone marrow-derived stromal cells. Reversine suppresses MCF-7 tumour growth and bone metastasis in vivo by reducing tumour stromalization including collagen deposition and recruitment of activated stromal cells. This study advocates μTSA as a platform for studying tumour microenvironmental interactions and cancer field effects with applications in drug discovery and development.
Conflict of interest statement
K.S. and B.P. are listed as inventors for a patent application disclosing μTSA, which includes data described in the manuscript.
Figures




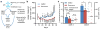
References
-
- Allinen M, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. - PubMed
-
- Gao MQ, et al. Stromal fibroblasts from the interface zone of human breast carcinomas induce an epithelial-mesenchymal transition-like state in breast cancer cells in vitro. J Cell Sci. 2010;123:3507–3514. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

