Synergy in protein-osmolyte mixtures
- PMID: 25490052
- PMCID: PMC4291039
- DOI: 10.1021/jp5111339
Synergy in protein-osmolyte mixtures
Abstract
Virtually all taxa use osmolytes to protect cells against biochemical stress. Osmolytes often occur in mixtures, such as the classical combination of urea with TMAO (trimethylamine N-oxide) in cartilaginous fish or the cocktail of at least six different osmolytes in the kidney. The concentration patterns of osmolyte mixtures found in vivo make it likely that synergy between them plays an important role. Using statistical mechanical n-component Kirkwood-Buff theory, we show from first principles that synergy in protein-osmolyte systems can arise from two separable sources: (1) mutual alteration of protein surface solvation and (2) effects mediated through bulk osmolyte chemical activities. We illustrate both effects in a four-component system with the experimental example of the unfolding of a notch ankyrin domain in urea-TMAO mixtures, which make urea a less effective denaturant and TMAO a more effective stabilizer. Protein surface effects are primarily responsible for this synergy. The specific patterns of surface solvation point to denatured state expansion as the main factor, as opposed to direct competition.
Figures

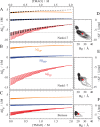
 UP, orange), TMAO (Δ
UP, orange), TMAO (Δ TP, red), and water (Δ
TP, red), and water (Δ WP, blue). The curves were calculated
using eqs 8 and 10. Each
group of curves represents increasing urea concentration from 0 to
3.5 M in 0.5 M steps (long to short curves). The dashed lines in panel
A indicate a solvation pattern that would result in a lack of synergy.
Panels D–F show an estimate for the change of TMAO excluded
volume upon unfolding vs the radius of gyration of the D state. Distributions/averages
for TMAO dihydrate are shown in black/red.
WP, blue). The curves were calculated
using eqs 8 and 10. Each
group of curves represents increasing urea concentration from 0 to
3.5 M in 0.5 M steps (long to short curves). The dashed lines in panel
A indicate a solvation pattern that would result in a lack of synergy.
Panels D–F show an estimate for the change of TMAO excluded
volume upon unfolding vs the radius of gyration of the D state. Distributions/averages
for TMAO dihydrate are shown in black/red.
Similar articles
-
Mutual Exclusion of Urea and Trimethylamine N-Oxide from Amino Acids in Mixed Solvent Environment.J Phys Chem Lett. 2015 Feb 19;6(4):581-5. doi: 10.1021/jz502634k. Epub 2015 Jan 28. J Phys Chem Lett. 2015. PMID: 26262470
-
Volume exclusion and H-bonding dominate the thermodynamics and solvation of trimethylamine-N-oxide in aqueous urea.J Am Chem Soc. 2012 Feb 22;134(7):3590-7. doi: 10.1021/ja211530n. Epub 2012 Feb 10. J Am Chem Soc. 2012. PMID: 22280147 Free PMC article.
-
Why do some organisms use a urea-methylamine mixture as osmolyte? Thermodynamic compensation of urea and trimethylamine N-oxide interactions with protein.Biochemistry. 1994 Oct 25;33(42):12695-701. doi: 10.1021/bi00208a021. Biochemistry. 1994. PMID: 7918496
-
Molecular basis of osmolyte effects on protein and metabolites.Methods Enzymol. 2007;428:459-86. doi: 10.1016/S0076-6879(07)28026-7. Methods Enzymol. 2007. PMID: 17875434 Review.
-
Living with urea stress.J Biosci. 2009 Jun;34(2):321-31. doi: 10.1007/s12038-009-0036-0. J Biosci. 2009. PMID: 19550048 Review.
Cited by
-
Deep Eutectic Solvents for Efficient Drug Solvation: Optimizing Composition and Ratio for Solubility of β-Cyclodextrin.Pharmaceutics. 2023 May 11;15(5):1462. doi: 10.3390/pharmaceutics15051462. Pharmaceutics. 2023. PMID: 37242704 Free PMC article.
-
Synergistic Solvation as the Enhancement of Local Mixing.J Phys Chem B. 2024 Jun 13;128(23):5713-5726. doi: 10.1021/acs.jpcb.4c01582. Epub 2024 Jun 3. J Phys Chem B. 2024. PMID: 38829987 Free PMC article.
-
Solvent accessible surface area-assessed molecular basis of osmolyte-induced protein stability.RSC Adv. 2024 Aug 9;14(34):25031-25041. doi: 10.1039/d4ra02576h. eCollection 2024 Aug 5. RSC Adv. 2024. PMID: 39131493 Free PMC article.
-
Combinations of Osmolytes, Including Monosaccharides, Disaccharides, and Sugar Alcohols Act in Concert During Cryopreservation to Improve Mesenchymal Stromal Cell Survival.Tissue Eng Part C Methods. 2016 Nov;22(11):999-1008. doi: 10.1089/ten.TEC.2016.0284. Epub 2016 Oct 27. Tissue Eng Part C Methods. 2016. PMID: 27758133 Free PMC article.
References
-
- Yancey P.; Clark M.; Hand S.; Bowlus R.; Somero G. Living with water stress: Evolution of osmolyte systems. Science 1982, 217, 1214–1222. - PubMed
-
- Greene R. J.; Pace C. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, α-chymotrypsin, and β-lactoglobulin. J. Biol. Chem. 1974, 249, 5388–5393. - PubMed
-
- Baskakov I.; Bolen D. W. Forcing thermodynamically unfolded proteins to fold. J. Biol. Chem. 1998, 273, 4831–4834. - PubMed
-
- Courtenay E.; Capp M.; Anderson C.; Record M. J. Vapor pressure osmometry studies of osmolyte-protein interactions: Implications for the action of osmoprotectants in vivo and for the interpretation of ”osmotic stress” experiments in vitro. Biochemistry 2000, 39, 4455–4471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

