Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing
- PMID: 25490068
- PMCID: PMC4270096
- DOI: 10.7554/eLife.04288
Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing
Erratum in
-
Correction: Spatial quality control bypasses cell-based limitations on proteostasis to promote prion curing.Elife. 2015 Jan 28;4:e06494. doi: 10.7554/eLife.06494. Elife. 2015. PMID: 25626954 Free PMC article. No abstract available.
Abstract
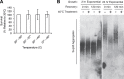
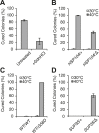
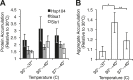
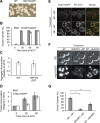
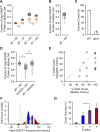
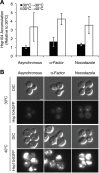
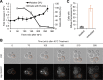
The proteostasis network has evolved to support protein folding under normal conditions and to expand this capacity in response to proteotoxic stresses. Nevertheless, many pathogenic states are associated with protein misfolding, revealing in vivo limitations on quality control mechanisms. One contributor to these limitations is the physical characteristics of misfolded proteins, as exemplified by amyloids, which are largely resistant to clearance. However, other limitations imposed by the cellular environment are poorly understood. To identify cell-based restrictions on proteostasis capacity, we determined the mechanism by which thermal stress cures the [PSI(+)]/Sup35 prion. Remarkably, Sup35 amyloid is disassembled at elevated temperatures by the molecular chaperone Hsp104. This process requires Hsp104 engagement with heat-induced non-prion aggregates in late cell-cycle stage cells, which promotes its asymmetric retention and thereby effective activity. Thus, cell division imposes a potent limitation on proteostasis capacity that can be bypassed by the spatial engagement of a quality control factor.
Keywords: S. cerevisiae; amyloid; cell biology; chaperone; prion; protein misfolding.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures






Similar articles
-
N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression.Genetics. 2006 Jun;173(2):611-20. doi: 10.1534/genetics.106.056820. Epub 2006 Apr 2. Genetics. 2006. PMID: 16582428 Free PMC article.
-
Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions.EMBO J. 2008 Oct 22;27(20):2712-24. doi: 10.1038/emboj.2008.194. Epub 2008 Oct 2. EMBO J. 2008. PMID: 18833196 Free PMC article.
-
The interaction of Hsp104 with yeast prion Sup35 as analyzed by fluorescence cross-correlation spectroscopy.Biochem Biophys Res Commun. 2013 Dec 6;442(1-2):28-32. doi: 10.1016/j.bbrc.2013.10.147. Epub 2013 Nov 8. Biochem Biophys Res Commun. 2013. PMID: 24216111
-
Prions, Chaperones, and Proteostasis in Yeast.Cold Spring Harb Perspect Biol. 2017 Feb 1;9(2):a023663. doi: 10.1101/cshperspect.a023663. Cold Spring Harb Perspect Biol. 2017. PMID: 27815300 Free PMC article. Review.
-
Structure and function of the molecular chaperone Hsp104 from yeast.Biopolymers. 2010 Mar;93(3):252-76. doi: 10.1002/bip.21301. Biopolymers. 2010. PMID: 19768774 Review.
Cited by
-
Hsp40 function in yeast prion propagation: Amyloid diversity necessitates chaperone functional complexity.Prion. 2015;9(2):80-9. doi: 10.1080/19336896.2015.1020268. Prion. 2015. PMID: 25738774 Free PMC article.
-
Beyond Amyloid Fibers: Accumulation, Biological Relevance, and Regulation of Higher-Order Prion Architectures.Viruses. 2022 Jul 27;14(8):1635. doi: 10.3390/v14081635. Viruses. 2022. PMID: 35893700 Free PMC article. Review.
-
[PIN+]ing down the mechanism of prion appearance.FEMS Yeast Res. 2018 May 1;18(3):foy026. doi: 10.1093/femsyr/foy026. FEMS Yeast Res. 2018. PMID: 29718197 Free PMC article. Review.
-
DMSO-mediated curing of several yeast prion variants involves Hsp104 expression and protein solubilization, and is decreased in several autophagy related gene (atg) mutants.PLoS One. 2020 Mar 5;15(3):e0229796. doi: 10.1371/journal.pone.0229796. eCollection 2020. PLoS One. 2020. PMID: 32134970 Free PMC article.
-
The [PSI +] yeast prion does not wildly affect proteome composition whereas selective pressure exerted on [PSI +] cells can promote aneuploidy.Sci Rep. 2017 Aug 16;7(1):8442. doi: 10.1038/s41598-017-07999-8. Sci Rep. 2017. PMID: 28814753 Free PMC article.
References
-
- Carmichael J, Chatellier J, Woolfson A, Milstein C, Fersht AR, Rubinsztein DC. Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington's disease. Proceedings of the National Academy of Sciences of USA. 2000;97:9701–9705. doi: 10.1073/pnas.170280697. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

