Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex
- PMID: 25490155
- PMCID: PMC4281882
- DOI: 10.7554/eLife.05115
Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex
Abstract
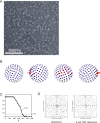
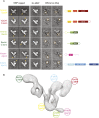
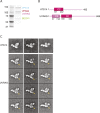
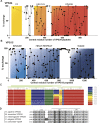
The class III phosphatidylinositol 3-kinase complex I (PI3KC3-C1) that functions in early autophagy consists of the lipid kinase VPS34, the scaffolding protein VPS15, the tumor suppressor BECN1, and the autophagy-specific subunit ATG14. The structure of the ATG14-containing PI3KC3-C1 was determined by single-particle EM, revealing a V-shaped architecture. All of the ordered domains of VPS34, VPS15, and BECN1 were mapped by MBP tagging. The dynamics of the complex were defined using hydrogen-deuterium exchange, revealing a novel 20-residue ordered region C-terminal to the VPS34 C2 domain. VPS15 organizes the complex and serves as a bridge between VPS34 and the ATG14:BECN1 subcomplex. Dynamic transitions occur in which the lipid kinase domain is ejected from the complex and VPS15 pivots at the base of the V. The N-terminus of BECN1, the target for signaling inputs, resides near the pivot point. These observations provide a framework for understanding the allosteric regulation of lipid kinase activity.
Keywords: autophagy; biophysics; human; hydrogen–deuterium exchange; lipid kinase; protein kinase; structural biology; three dimensional electron microscopy.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

