chromoWIZ: a web tool to query and visualize chromosome-anchored genes from cereal and model genomes
- PMID: 25491094
- PMCID: PMC4266971
- DOI: 10.1186/s12870-014-0348-6
chromoWIZ: a web tool to query and visualize chromosome-anchored genes from cereal and model genomes
Abstract
Background: Over the last years reference genome sequences of several economically and scientifically important cereals and model plants became available. Despite the agricultural significance of these crops only a small number of tools exist that allow users to inspect and visualize the genomic position of genes of interest in an interactive manner.
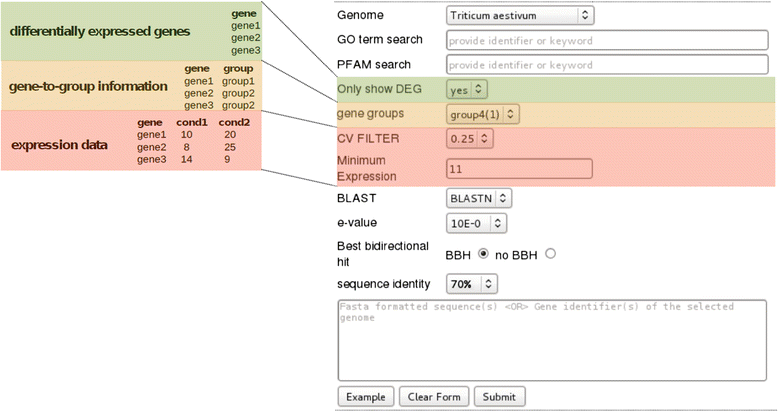
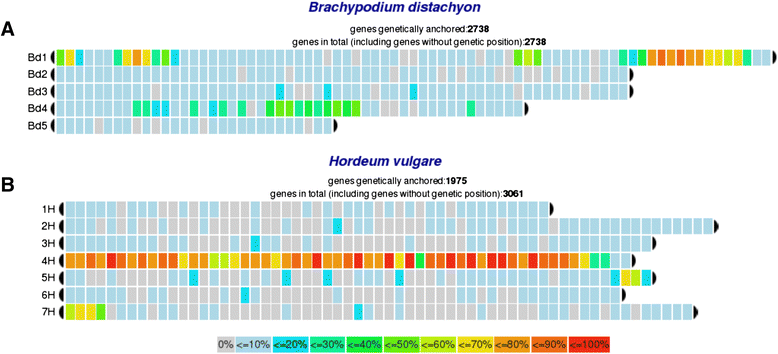
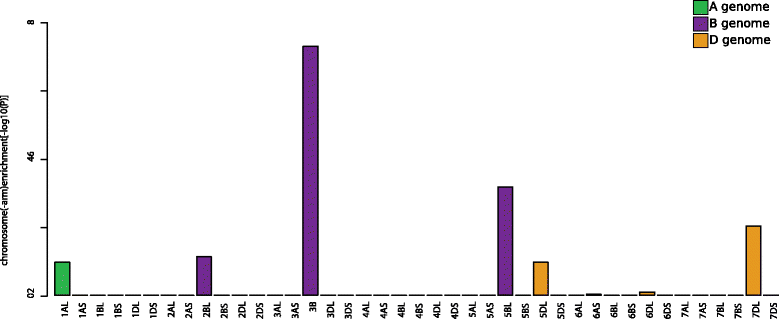
Description: We present chromoWIZ, a web tool that allows visualizing the genomic positions of relevant genes and comparing these data between different plant genomes. Genes can be queried using gene identifiers, functional annotations, or sequence homology in four grass species (Triticum aestivum, Hordeum vulgare, Brachypodium distachyon, Oryza sativa). The distribution of the anchored genes is visualized along the chromosomes by using heat maps. Custom gene expression measurements, differential expression information, and gene-to-group mappings can be uploaded and can be used for further filtering.
Conclusions: This tool is mainly designed for breeders and plant researchers, who are interested in the location and the distribution of candidate genes as well as in the syntenic relationships between different grass species. chromoWIZ is freely available and online accessible at http://mips.helmholtz-muenchen.de/plant/chromoWIZ/index.jsp.
Figures

References
-
- Michael TP, Jackson S: The first 50 plant genomes.Plant Genome 2013, 6(2). https://www.crops.org/publications/tpg/articles/6/2/plantgenome2013.03.0....
-
- Morrell PL, Buckler ES, Ross-Ibarra J. Crop genomics: advances and applications. Nat Rev Genet. 2011;13(2):85–96. - PubMed
-
- Mayer KF, Martis M, Hedley PE, Simkova H, Liu H, Morris JA, Steuernagel B, Taudien S, Roessner S, Gundlach H, Kubalakova M, Suchankova P, Murat F, Felder M, Nussbaumer T, Graner A, Salse J, Endo T, Sakai H, Tanaka T, Itoh T, Sato K, Platzer M, Matsumoto T, Scholz U, Dolezel J, Waugh R, Stein N. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell. 2011;23(4):1249–1263. doi: 10.1105/tpc.110.082537. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

