Two different pathways of phosphatidylcholine synthesis, the Kennedy Pathway and the Lands Cycle, differentially regulate cellular triacylglycerol storage
- PMID: 25491198
- PMCID: PMC4293825
- DOI: 10.1186/s12860-014-0043-3
Two different pathways of phosphatidylcholine synthesis, the Kennedy Pathway and the Lands Cycle, differentially regulate cellular triacylglycerol storage
Abstract
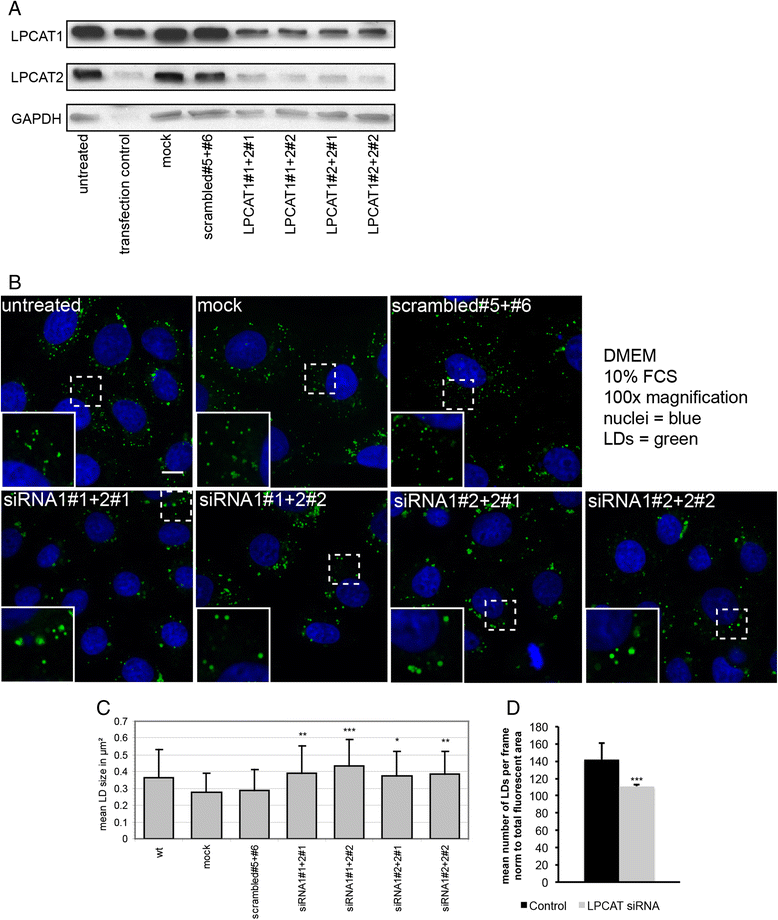
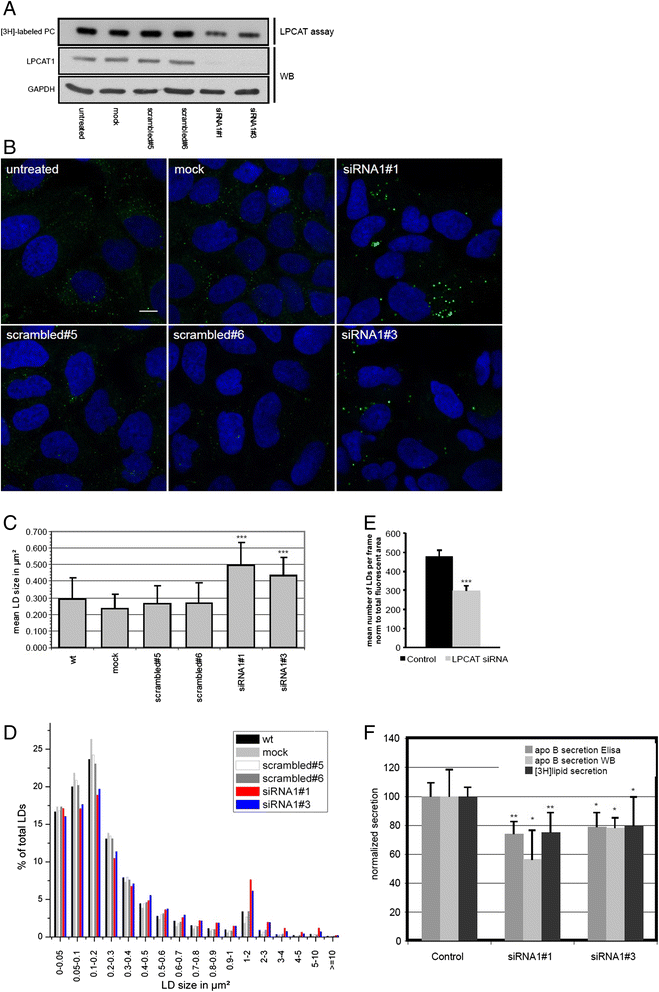
Background: Lipids are stored within cells in lipid droplets (LDs). They consist of a core of neutral lipids surrounded by a monolayer of phospholipids, predominantly phosphatidylcholine (PC). LDs are very dynamic and can rapidly change in size upon lipid uptake or release. These dynamics require a fast adaptation of LD surface. We have recently shown that two Lands cycle PC synthesizing enyzmes, LPCAT1 and LPCAT2 can localize to the LD surface.
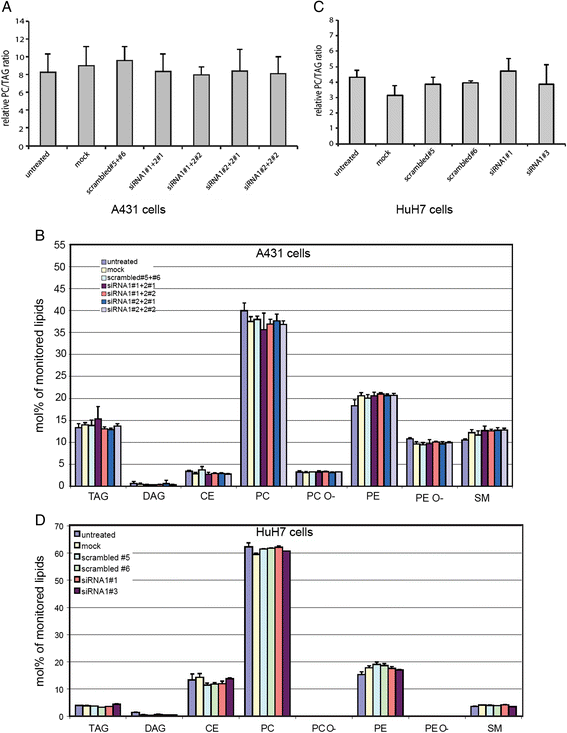
Results: Here, we show that knock-down of both enzymes leads to an increase in LD size without changes in the total amount of neutral lipids, while interference with the de-novo Kennedy pathway PC biosynthesis is associated with changes in triacylglyceride synthesis. We show that function of LPCAT1 and 2 is conserved in Drosophila melanogaster by the ortholog CG32699. Furthermore we demonstrate that modulation of the LD pool by LPCAT1 influences the release of lipoprotein from liver cells.
Conclusion: Activity of the Kennedy pathway regulates the balance between phospholipids and neutral lipids, while the Lands cycle regulates lipid droplet size by regulating surface availability and influencing surface to volume ratio. Differences in lipid droplet size may account for differences in lipid dynamics and be relevant to understand lipid overload diseases.
Figures



References
-
- Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644(1):47–59. doi: 10.1016/j.bbamcr.2003.10.018. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

