Acute-phase protein serum amyloid A3 is a novel paracrine coupling factor that controls bone homeostasis
- PMID: 25491310
- PMCID: PMC4396610
- DOI: 10.1096/fj.14-265512
Acute-phase protein serum amyloid A3 is a novel paracrine coupling factor that controls bone homeostasis
Abstract
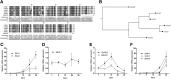
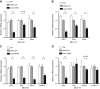
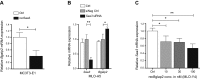
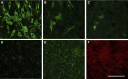
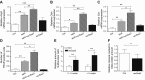
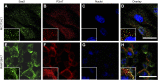
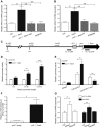
Serum amyloid A (A-SAA/Saa3) was shown before to affect osteoblastic metabolism. Here, using RT-quantitative PCR and/or immunoblotting, we show that expression of mouse Saa3 and human SAA1 and SAA2 positively correlates with increased cellular maturation toward the osteocyte phenotype. Expression is not detected in C3H10T1/2 embryonic fibroblasts but is successively higher in preosteoblastic MC3T3-E1 cells, late osteoblastic MLO-A5 cells, and MLO-Y4 osteocytes, consistent with findings using primary bone cells from newborn mouse calvaria. Recombinant Saa3 protein functionally inhibits osteoblast differentiation as reflected by reductions in the expression of osteoblast markers and decreased mineralization in newborn mouse calvaria. Yet, Saa3 protein enhances osteoclastogenesis in mouse macrophages/monocytes based on the number of multinucleated and tartrate-resistant alkaline phosphatase-positive cells and Calcr mRNA expression. Depletion of Saa3 in MLO osteocytes results in the loss of the mature osteocyte phenotype. Recombinant osteocalcin, which is reciprocally regulated with Saa3 at the osteoblast/osteocyte transition, attenuates Saa3 expression in MLO-Y4 osteocytes. Mechanistically, Saa3 produced by MLO-Y4 osteocytes is integrated into the extracellular matrix of MC3T3-E1 osteoblasts, where it associates with the P2 purinergic receptor P2rx7 to stimulate Mmp13 expression via the P2rx7/MAPK/ERK/activator protein 1 axis. Our data suggest that Saa3 may function as an important coupling factor in bone development and homeostasis.
Keywords: osteoblast; osteoclast; osteocyte; osteogenesis.
© FASEB.
Figures

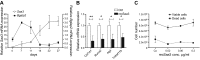
Similar articles
-
IL-6 Enhances Osteocyte-Mediated Osteoclastogenesis by Promoting JAK2 and RANKL Activity In Vitro.Cell Physiol Biochem. 2017;41(4):1360-1369. doi: 10.1159/000465455. Epub 2017 Mar 9. Cell Physiol Biochem. 2017. PMID: 28278513
-
MLO-Y4 osteocyte-like cells support osteoclast formation and activation.J Bone Miner Res. 2002 Nov;17(11):2068-79. doi: 10.1359/jbmr.2002.17.11.2068. J Bone Miner Res. 2002. PMID: 12412815
-
Oxygen tension is an important mediator of the transformation of osteoblasts to osteocytes.J Bone Miner Metab. 2007;25(5):266-76. doi: 10.1007/s00774-007-0765-9. Epub 2007 Aug 25. J Bone Miner Metab. 2007. PMID: 17704991
-
The local production of 1,25(OH)2D3 promotes osteoblast and osteocyte maturation.J Steroid Biochem Mol Biol. 2014 Oct;144 Pt A:114-8. doi: 10.1016/j.jsbmb.2013.10.003. Epub 2013 Oct 12. J Steroid Biochem Mol Biol. 2014. PMID: 24125735 Review.
-
Role of Galectin-3 in Bone Cell Differentiation, Bone Pathophysiology and Vascular Osteogenesis.Int J Mol Sci. 2017 Nov 21;18(11):2481. doi: 10.3390/ijms18112481. Int J Mol Sci. 2017. PMID: 29160796 Free PMC article. Review.
Cited by
-
The White-Spotted Bamboo Shark Genome Reveals Chromosome Rearrangements and Fast-Evolving Immune Genes of Cartilaginous Fish.iScience. 2020 Nov 2;23(11):101754. doi: 10.1016/j.isci.2020.101754. eCollection 2020 Nov 20. iScience. 2020. PMID: 33251490 Free PMC article.
-
Infrapatellar Fat Pad and Knee Osteoarthritis.Aging Dis. 2020 Oct 1;11(5):1317-1328. doi: 10.14336/AD.2019.1116. eCollection 2020 Oct. Aging Dis. 2020. PMID: 33014539 Free PMC article. Review.
-
Akkermansia muciniphila promotes type H vessel formation and bone fracture healing by reducing gut permeability and inflammation.Dis Model Mech. 2020 Dec 1;13(11):dmm043620. doi: 10.1242/dmm.043620. Dis Model Mech. 2020. PMID: 33033107 Free PMC article.
-
Nano-Hydroxyapatite Stimulation of Gene Expression Requires Fgf Receptor, Phosphate Transporter, and Erk1/2 Signaling.ACS Appl Mater Interfaces. 2017 Nov 15;9(45):39185-39196. doi: 10.1021/acsami.7b12029. Epub 2017 Oct 31. ACS Appl Mater Interfaces. 2017. PMID: 29045789 Free PMC article.
-
Nfib promotes chondrocyte proliferation and inhibits differentiation by mildly regulating Sox9 and its downstream genes.Mol Biol Rep. 2021 Nov;48(11):7487-7497. doi: 10.1007/s11033-021-06767-2. Epub 2021 Oct 15. Mol Biol Rep. 2021. PMID: 34651294
References
-
- Li X., Cao X. (2006) BMP signaling and skeletogenesis. Ann. N. Y. Acad. Sci. 1068, 26–40 - PubMed
-
- Hadjidakis D. J., Androulakis I. I. (2006) Bone remodeling. Ann. N. Y. Acad. Sci. 1092, 385–396 - PubMed
-
- Kong Y. Y., Yoshida H., Sarosi I., Tan H. L., Timms E., Capparelli C., Morony S., Oliveira-dos-Santos A. J., Van G., Itie A., Khoo W., Wakeham A., Dunstan C. R., Lacey D. L., Mak T. W., Boyle W. J., Penninger J. M. (1999) OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 397, 315–323 - PubMed
-
- Del Fattore A., Teti A., Rucci N. (2012) Bone cells and the mechanisms of bone remodelling. Front Biosci. (Elite Ed.) 4, 2302–2321 - PubMed
-
- Matsuoka K., Park K. A., Ito M., Ikeda K., Takeshita S. (2014) Osteoclast-derived complement component 3a stimulates osteoblast differentiation. J. Bone Miner. Res. 29, 1522–1530 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

