Sequencing and phylogenetic analysis of near full-length HIV-1 subtypes A, B, G and unique recombinant AC and AD viral strains identified in South Africa
- PMID: 25492033
- PMCID: PMC4378615
- DOI: 10.1089/AID.2014.0230
Sequencing and phylogenetic analysis of near full-length HIV-1 subtypes A, B, G and unique recombinant AC and AD viral strains identified in South Africa
Abstract
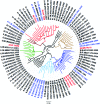
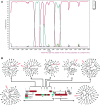
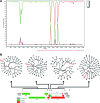
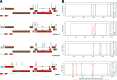
By the end of 2012, more than 6.1 million people were infected with HIV-1 in South Africa. Subtype C was responsible for the majority of these infections and more than 300 near full-length genomes (NFLGs) have been published. Currently very few non-subtype C isolates have been identified and characterized within the country, particularly full genome non-C isolates. Seven patients from the Tygerberg Virology (TV) cohort were previously identified as possible non-C subtypes and were selected for further analyses. RNA was isolated from five individuals (TV047, TV096, TV101, TV218, and TV546) and DNA from TV016 and TV1057. The NFLGs of these samples were amplified in overlapping fragments and sequenced. Online subtyping tools REGA version 3 and jpHMM were used to screen for subtypes and recombinants. Maximum likelihood (ML) phylogenetic analysis (phyML) was used to infer subtypes and SimPlot was used to confirm possible intersubtype recombinants. We identified three subtype B (TV016, TV047, and TV1057) isolates, one subtype A1 (TV096), one subtype G (TV546), one unique AD (TV101), and one unique AC (TV218) recombinant form. This is the first NFLG of subtype G that has been described in South Africa. The subtype B sequences described also increased the NFLG subtype B sequences in Africa from three to six. There is a need for more NFLG sequences, as partial HIV-1 sequences may underrepresent viral recombinant forms. It is also necessary to continue monitoring the evolution and spread of HIV-1 in South Africa, because understanding viral diversity may play an important role in HIV-1 prevention strategies.
Figures
Similar articles
-
Molecular characterization of non-subtype C and recombinant HIV-1 viruses from Cape Town, South Africa.Infect Genet Evol. 2009 Sep;9(5):840-6. doi: 10.1016/j.meegid.2009.05.001. Epub 2009 May 14. Infect Genet Evol. 2009. PMID: 19446657
-
Characterization and frequency of a newly identified HIV-1 BF1 intersubtype circulating recombinant form in São Paulo, Brazil.Virol J. 2010 Apr 16;7:74. doi: 10.1186/1743-422X-7-74. Virol J. 2010. PMID: 20398371 Free PMC article.
-
Identification of new CRF43_02G and CRF25_cpx in Saudi Arabia based on full genome sequence analysis of six HIV type 1 isolates.AIDS Res Hum Retroviruses. 2008 Oct;24(10):1327-35. doi: 10.1089/aid.2008.0101. AIDS Res Hum Retroviruses. 2008. PMID: 18844465
-
HIV subtype diversity worldwide.Curr Opin HIV AIDS. 2019 May;14(3):153-160. doi: 10.1097/COH.0000000000000534. Curr Opin HIV AIDS. 2019. PMID: 30882484 Review.
-
Evolution and diversity of HIV-1 in Africa--a review.Virus Genes. 2003;26(2):151-63. doi: 10.1023/a:1023435429841. Virus Genes. 2003. PMID: 12803467 Review.
Cited by
-
Deep sequencing of near full-length HIV-1 genomes from plasma identifies circulating subtype C and infrequent occurrence of AC recombinant form in Southern India.PLoS One. 2017 Dec 8;12(12):e0188603. doi: 10.1371/journal.pone.0188603. eCollection 2017. PLoS One. 2017. PMID: 29220350 Free PMC article.
-
Molecular Epidemiology and Trends in HIV-1 Transmitted Drug Resistance in Mozambique 1999-2018.Viruses. 2022 Sep 9;14(9):1992. doi: 10.3390/v14091992. Viruses. 2022. PMID: 36146798 Free PMC article.
-
Molecular Epidemiology of HIV-1 Subtype G in the Russian Federation.Viruses. 2019 Apr 16;11(4):348. doi: 10.3390/v11040348. Viruses. 2019. PMID: 30995717 Free PMC article.
-
Near full-length HIV-1 subtype B sequences from the early South African epidemic, detecting a BD unique recombinant form (URF) from a sample in 1985.Sci Rep. 2019 Apr 17;9(1):6227. doi: 10.1038/s41598-019-42417-1. Sci Rep. 2019. PMID: 30996293 Free PMC article.
-
Validation of Variant Assembly Using HAPHPIPE with Next-Generation Sequence Data from Viruses.Viruses. 2020 Jul 14;12(7):758. doi: 10.3390/v12070758. Viruses. 2020. PMID: 32674515 Free PMC article.
References
-
- Shisana O, Rehle T, Simbayi LC, et al. : South African National HIV Prevalence, Incidence and Behaviour Survey, 2012. HSRC Press, Cape Town
-
- Hemelaar J: Implications of HIV diversity for the HIV-1 pandemic. J Infect Dis 2013;66(5):391–400 - PubMed
-
- van Harmelen J, Williamson C, Kim B, et al. : Characterization of full-length HIV type 1 subtype C sequences from South Africa. AIDS Res Hum Retroviruses 2001;17(16):1527–1531 - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
