Fragmentation of tRNA in Phytophthora infestans asexual life cycle stages and during host plant infection
- PMID: 25492044
- PMCID: PMC4272539
- DOI: 10.1186/s12866-014-0308-1
Fragmentation of tRNA in Phytophthora infestans asexual life cycle stages and during host plant infection
Abstract
Background: The oomycete Phytophthora infestans possesses active RNA silencing pathways, which presumably enable this plant pathogen to control the large numbers of transposable elements present in its 240 Mb genome. Small RNAs (sRNAs), central molecules in RNA silencing, are known to also play key roles in this organism, notably in regulation of critical effector genes needed for infection of its potato host.
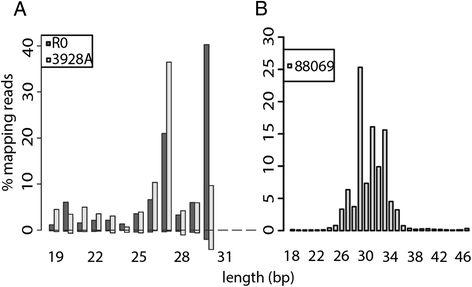
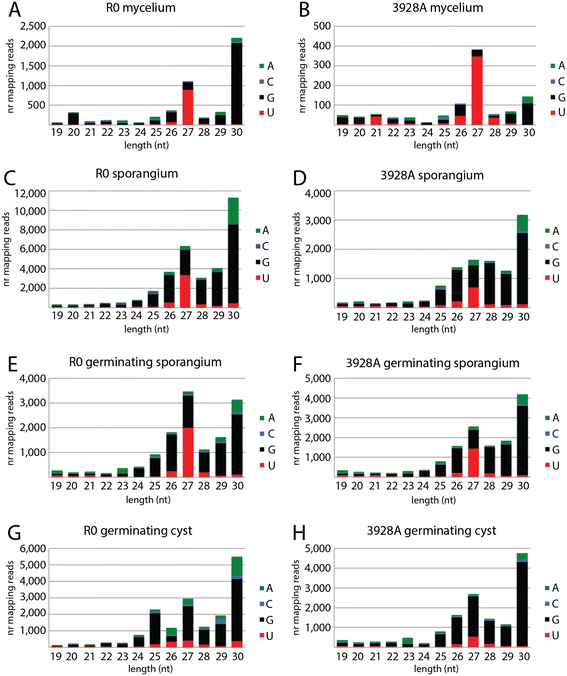
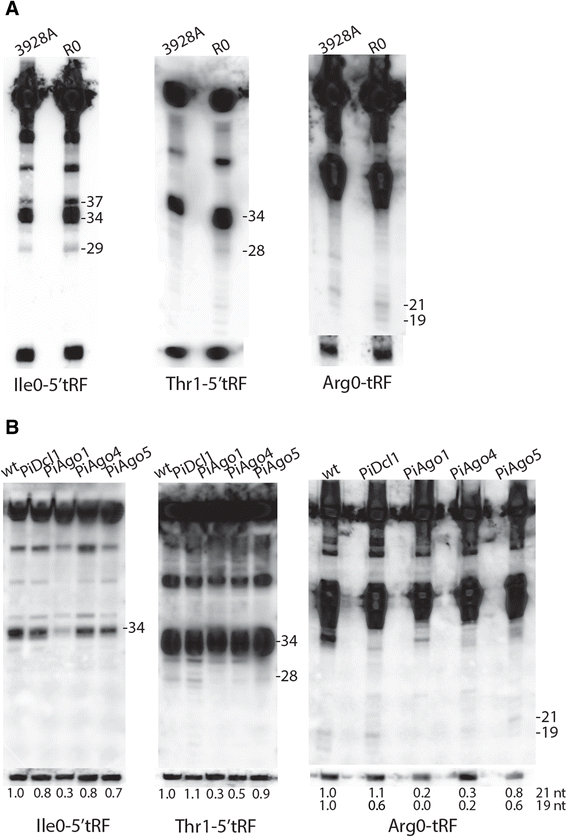
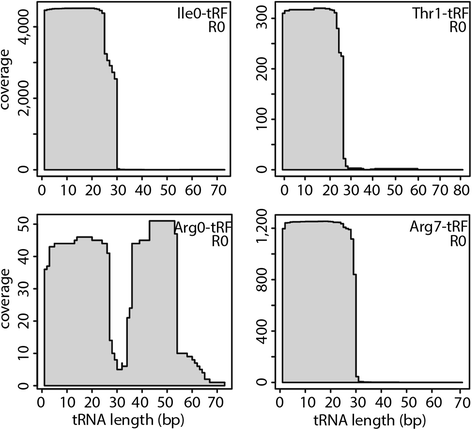
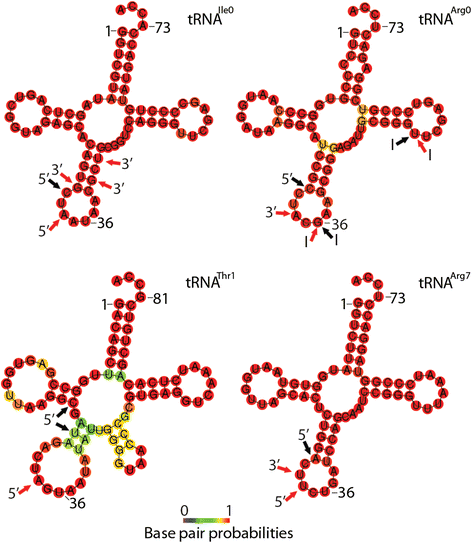
Results: To identify additional classes of sRNAs in oomycetes, we mapped deep sequencing reads to transfer RNAs (tRNAs) thereby revealing the presence of 19-40 nt tRNA-derived RNA fragments (tRFs). Northern blot analysis identified abundant tRFs corresponding to half tRNA molecules. Some tRFs accumulated differentially during infection, as seen by examining sRNAs sequenced from P. infestans-potato interaction libraries. The putative connection between tRF biogenesis and the canonical RNA silencing pathways was investigated by employing hairpin RNA-mediated RNAi to silence the genes encoding P. infestans Argonaute (PiAgo) and Dicer (PiDcl) endoribonucleases. By sRNA sequencing we show that tRF accumulation is PiDcl1-independent, while Northern hybridizations detected reduced levels of specific tRNA-derived species in the PiAgo1 knockdown line.
Conclusions: Our findings extend the sRNA diversity in oomycetes to include fragments derived from non-protein-coding RNA transcripts and identify tRFs with elevated levels during infection of potato by P. infestans.
Figures
Similar articles
-
Evidence for small RNAs homologous to effector-encoding genes and transposable elements in the oomycete Phytophthora infestans.PLoS One. 2012;7(12):e51399. doi: 10.1371/journal.pone.0051399. Epub 2012 Dec 14. PLoS One. 2012. PMID: 23272103 Free PMC article.
-
Phytophthora infestans Argonaute 1 binds microRNA and small RNAs from effector genes and transposable elements.New Phytol. 2016 Aug;211(3):993-1007. doi: 10.1111/nph.13946. Epub 2016 Mar 24. New Phytol. 2016. PMID: 27010746
-
Evidence for involvement of Dicer-like, Argonaute and histone deacetylase proteins in gene silencing in Phytophthora infestans.Mol Plant Pathol. 2011 Oct;12(8):772-85. doi: 10.1111/j.1364-3703.2011.00710.x. Epub 2011 Mar 29. Mol Plant Pathol. 2011. PMID: 21726377 Free PMC article.
-
How Does Phytophthora infestans Evade Control Efforts? Modern Insight Into the Late Blight Disease.Phytopathology. 2018 Aug;108(8):916-924. doi: 10.1094/PHYTO-04-18-0130-IA. Epub 2018 Jul 6. Phytopathology. 2018. PMID: 29979126 Review.
-
Five Reasons to Consider Phytophthora infestans a Reemerging Pathogen.Phytopathology. 2015 Jul;105(7):966-81. doi: 10.1094/PHYTO-01-15-0005-FI. Epub 2015 Jun 26. Phytopathology. 2015. PMID: 25760519 Review.
Cited by
-
tRNA-Derived Fragments (tRFs): Emerging New Roles for an Ancient RNA in the Regulation of Gene Expression.Life (Basel). 2015 Nov 27;5(4):1638-51. doi: 10.3390/life5041638. Life (Basel). 2015. PMID: 26703738 Free PMC article. Review.
-
Fine-Tuning of Gene Expression by tRNA-Derived Fragments during Abiotic Stress Signal Transduction.Int J Mol Sci. 2018 Feb 8;19(2):518. doi: 10.3390/ijms19020518. Int J Mol Sci. 2018. PMID: 29419808 Free PMC article. Review.
-
Computational Approaches to tRNA-Derived Small RNAs.Noncoding RNA. 2017 Jan 4;3(1):2. doi: 10.3390/ncrna3010002. Noncoding RNA. 2017. PMID: 29657274 Free PMC article. Review.
-
A novel and ubiquitous miRNA-involved regulatory module ensures precise phosphorylation of RNA polymerase II and proper transcription.PLoS Pathog. 2024 Apr 19;20(4):e1012138. doi: 10.1371/journal.ppat.1012138. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38640110 Free PMC article.
-
smartPARE: An R Package for Efficient Identification of True mRNA Cleavage Sites.Int J Mol Sci. 2021 Apr 20;22(8):4267. doi: 10.3390/ijms22084267. Int J Mol Sci. 2021. PMID: 33924042 Free PMC article.
References
-
- Dick MW. The Peronosporomycetes. In: McLaughlin DJ, McLaughlin EG, Lemke PA, editors. The Mycota VII. Systematics and Evolution Part A. Berlin Heidelberg New York: Springer; 2001. pp. 39–72.
-
- Adl SM, Simpson AG, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, Le Gall L, Lynn DH, McManus H, Mitchell EA, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick RS, Schoch CL, Smirnov A, Spiegel FW. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59(5):429–493. doi: 10.1111/j.1550-7408.2012.00644.x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

