Biochemical Properties and Biological Functions of FET Proteins
- PMID: 25494299
- PMCID: PMC9188303
- DOI: 10.1146/annurev-biochem-060614-034325
Biochemical Properties and Biological Functions of FET Proteins
Abstract
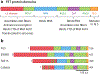
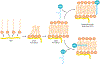
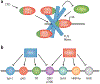
Members of the FET protein family, consisting of FUS, EWSR1, and TAF15, bind to RNA and contribute to the control of transcription, RNA processing, and the cytoplasmic fates of messenger RNAs in metazoa. FET proteins can also bind DNA, which may be important in transcription and DNA damage responses. FET proteins are of medical interest because chromosomal rearrangements of their genes promote various sarcomas and because point mutations in FUS or TAF15 can cause neurodegenerative diseases such as amyotrophic lateral sclerosis and frontotemporal lobar dementia. Recent results suggest that both the normal and pathological effects of FET proteins are modulated by low-complexity or prion-like domains, which can form higher-order assemblies with novel interaction properties. Herein, we review FET proteins with an emphasis on how the biochemical properties of FET proteins may relate to their biological functions and to pathogenesis.
Keywords: EWSR1; FUS; RNA-binding; TAF15; low-complexity; neurodegeneration.
Figures

References
-
- Eberle AB, Visa N. 2014. Quality control of mRNP biogenesis: networking at the transcription site. Semin. Cell Dev. Biol 32C:37–46 - PubMed
-
- Saguez C, Olesen JR, Jensen TH. 2005. Formation of export-competent mRNP: escaping nuclear destruction. Curr. Opin. Cell Biol 17:287–93 - PubMed
-
- Moore MJ, Proudfoot NJ. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136:688–700 - PubMed
-
- Hilleren P, Parker R. 1999. Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet 33:229–60 - PubMed
-
- Mitchell SF, Parker R. 2014. Principles and properties of eukaryotic mRNPs. Mol. Cell 54:547–58 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

