Ion-specific control of the self-assembly dynamics of a nanostructured protein lattice
- PMID: 25494454
- PMCID: PMC4310639
- DOI: 10.1021/nn502992x
Ion-specific control of the self-assembly dynamics of a nanostructured protein lattice
Abstract
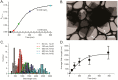
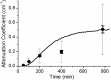
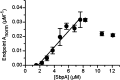
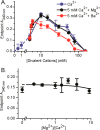
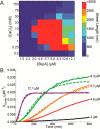
Self-assembling proteins offer a potential means of creating nanostructures with complex structure and function. However, using self-assembly to create nanostructures with long-range order whose size is tunable is challenging, because the kinetics and thermodynamics of protein interactions depend sensitively on solution conditions. Here we systematically investigate the impact of varying solution conditions on the self-assembly of SbpA, a surface-layer protein from Lysinibacillus sphaericus that forms two-dimensional nanosheets. Using high-throughput light scattering measurements, we mapped out diagrams that reveal the relative yield of self-assembly of nanosheets over a wide range of concentrations of SbpA and Ca(2+). These diagrams revealed a localized region of optimum yield of nanosheets at intermediate Ca(2+) concentration. Replacement of Mg(2+) or Ba(2+) for Ca(2+) indicates that Ca(2+) acts both as a specific ion that is required to induce self-assembly and as a general divalent cation. In addition, we use competitive titration experiments to find that 5 Ca(2+) bind to SbpA with an affinity of 67.1 ± 0.3 μM. Finally, we show via modeling that nanosheet assembly occurs by growth from a negligibly small critical nucleus. We also chart the dynamics of nanosheet size over a variety of conditions. Our results demonstrate control of the dynamics and size of the self-assembly of a nanostructured lattice, the constituents of which are one of a class of building blocks able to form novel hybrid nanomaterials.
Keywords: Ca2+ binding; biomaterials; nanostructures; protein interactions; self-assembly dynamics.
Figures


References
-
- Pavkov-Keller T.; Howorka S.; Keller W. The Structure of Bacterial S-Layer Proteins. Prog. Mol. Biol. Transl. Sci. 2011, 103, 73–130. - PubMed
-
- Vekilov P. G. The Two-Step Mechanism of Nucleation of Crystals in Solution. Nanoscale 2010, 2, 2346–2357. - PubMed
-
- Whitelam S. Control of Pathways and Yields of Protein Crystallization through the Interplay of Nonspecific and Specific Attractions. Phys. Rev. Lett. 2010, 105, 088102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

